 PDF(625 KB)
PDF(625 KB)


新世纪中国菌物新名称发表概况(2000-2020)
王科,陈双林,戴玉成,贾泽峰,李泰辉,刘铁志,普布多吉,热衣木·马木提,孙广宇,图力古尔,魏生龙,杨祝良,袁海生,张修国,蔡磊
菌物学报 ›› 2021, Vol. 40 ›› Issue (4) : 822-833.
 PDF(625 KB)
PDF(625 KB)
 PDF(625 KB)
PDF(625 KB)
新世纪中国菌物新名称发表概况(2000-2020)
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Overview of China’s nomenclature novelties of fungi in the new century (2000-2020)
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}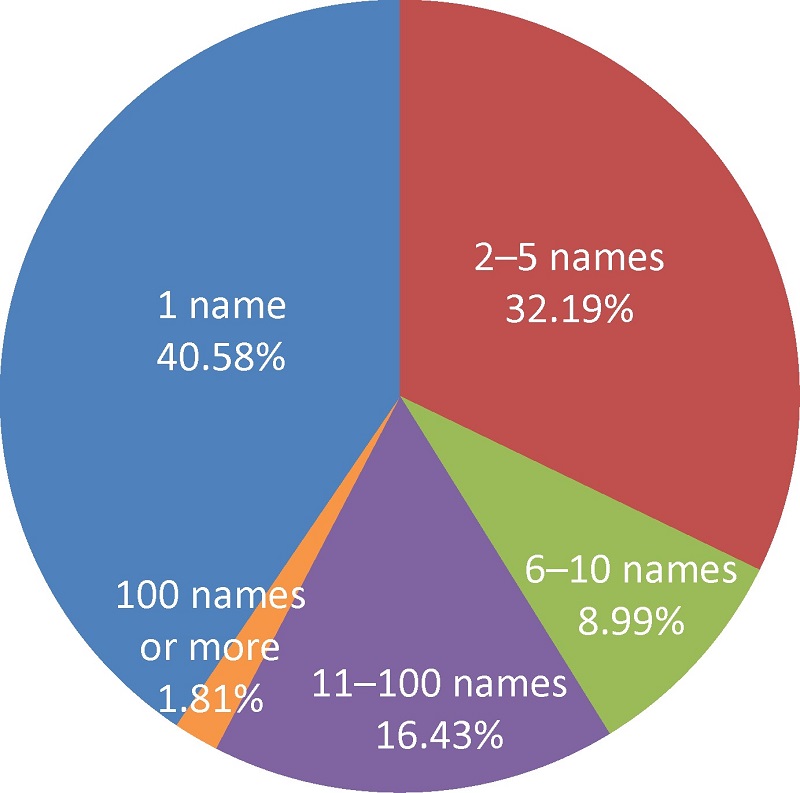
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |