INTRODUCTION
Species of the Gyromitra Fr. (Discinaceae, Pezizales), known as “false morels”, are impressed by producing cerebriform, saddle-shaped or discoid ascocarps (Methven et al. 2013; Wang & Zhuang 2019). At present, ca. 70 species were described and accepted worldwide (Kirk et al. 2008; Methven et al. 2013; Krisai-Greilhuber et al. 2017; Crous et al. 2018; Wang & Zhuang 2019;
Due to the morphological characteristics, six subgenera were recognized in Gyromitra, namely Caroliniana S.P. Abbott, Discina (Fr.) Harmaja, Gyromitra (Pers.) Fr., Melaleucoides S.P. Abbott, Pseudorhizina Methven, Zelski, and A.N. Mill., and Pseudoverpa P.A. Moreau, Bellanger & Loizides (Harmaja 1973; Abbott & Currah 1997; van Vooren & Moreau 2009a; Methven et al. 2013; Crous et al. 2018; Wang & Zhuang 2019), and some of the subgenera were further divided into different sections (van Vooren & Moreau 2009b, 2009c , 2009d). Subgenera Caroliniana and Discina share uniguttulate or triguttulate and apiculate ascospores, but the former has reticulate spores with multiple apiculi at each end compared to rugose spores with solitary apiculi at each end of the latter. In subgenus Melaleucoides, ascospores are biguttulate, nonapiculate, and smooth to verrucose. In subgenus Gyromitra, ascospores are biguttulate, nonapiculate or with a broadly rounded apiculus at each end, cyanophilic, smooth, or finely verrucose. In subgenus Pseudorhizina, ascospores are non-apiculate, acyanophilic, smooth, or finely rugose. In subgenus Pseudoverpa, ascospores are mostly biguttulate, non-apiculate, cyanophilic, and smooth.
Gyromitra species containing toxins gyromitrins have been authenticated to be capable of causing poisoning incidents worldwide and even deaths. Owing to the morphological similarities, species of Gyromitra were often mistaken as taxa of Morchella Dill. ex Pers. (Giusti & Carnevale 1974; Michelot & Toth 1991; Leathem & Dorran 2007; Patocka et al. 2012; Arłukowicz-Grabowska et al. 2019). Several species including G. ambigua (P. Karst.) Harmaja, G. esculenta (Pers.) Fr., G. fastigiata (Krombh.) Rehm, G. gigas (Krombh.) Cooke, G. infula (Schaeff.) Quél, and G. splendida Raitv. were demonstrated as poisonous, and G. esculenta was one of the most common species discovered from mushroom poisoning incidents (Michelot & Toth 1991; Mao 2006; Leathem & Dorran 2007; Patocka et al. 2012; Bau et al. 2014; Chen et al. 2014, 2016; Arłukowicz-Grabowska et al. 2019; Wu et al. 2019).
In March 2020, four people from Guizhou and Yunnan provinces were sent to emergency departments of local hospitals after consumption of some “morels”. Soon after the poisoning incidents, specimens were collected from the same localities and confirmed by the patients. Morphological and phylogenetic analyses indicated that the “morels” were identical to the specimens (MHHNU 7671, HKAS 107322) previously identified as “G. esculenta” from Hunan Province (Chen et al. 2016). Further studies showed that those specimens from China were a different taxon which could not match any known Gyromitra species. In the present study, it is described and illustrated as a new species.
1 MATERIALS AND METHODS
1.1 Sample collection and morphological study
The studied specimens were processed and deposited in the Cryptogamic Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (HKAS), the Herbarium of Mycology, Hunan Normal University (MHHNU), the Herbarium Mycologicum Academiae Sinicae (HMAS), and National Institute of Occupational Health and Poison Control (NIOHP), Chinese Center for Disease Control and Prevention. Observations on micro-morphological structures were carried out based on dried specimens following the method of Wang & Zhuang (2019). Sections were studied at a magnification of 1 000× with a Nikon E 80i microscope and phase contrast illumination. As described by the method of Wu et al. (2014, 2016), scanning electron microscope (SEM) was used for observing spore ornamentations. For description of ascospores, the abbreviation [n/m/p] means n ascospores measured from m apothecia of p collections; dimensions for ascospores are given as (a-)b-c(-d), wherein the range b-c contains a minimum of 90% of the measured values, extreme values (a, b) are given in parentheses. Q is used to express the “length/width ratio” of a spore in side view; Qm means the average Q of all ascospores measured ± sample standard deviation.
1.2 Molecular study and phylogenetic analyses
Protocols for DNA extraction, PCR, sequencing and sequence alignment followed those in Wu et al. (2014) and Wang & Zhuang (2019). Three DNA gene fragments including the internal transcribed spacer (ITS), the D1-D3 domains of the nuclear large subunit ribosomal DNA (nrLSU), and the translation elongation factor subunit 1α (tef1-α) were amplified. The primer pairs were ITS1F/ITS4 or ITS5/ITS4 (White et al. 1990; Gardes & Bruns 1993), LROR/LR5 (Vilgalys & Hester 1990), and TEF1-983 F/TEF1-1567R (Rehner & Buckley 2005), respectively.
Besides the sequences obtained from this study (GenBank nos. MT424855-MT424856, MT421931-MT421932, MT424599-MT424600, MT509846-MT509847, MT512574-MT512575, MT522867-MT522868), sequences of Gyromitra species were selected for phylogenetic analysis mainly according to previously studies (Methven et al. 2013; Crous et al. 2018; Wang & Zhuang 2019) and outgroups were selected by following Wang & Zhuang (2019). Likewise, two datasets, the ITS sequence matrix and a concatenated dataset including all three genes, were also compiled.
All sequences were aligned using ClustalX 1.83 (Chenna et al. 2003) and manually edited on BioEdit v7.0.5 (Hall 1999). Two independent methods, Maximum Likelihood analysis (ML) and Bayesian Inference (BI), were used for phylogenetic analyses, which were conducted on RAxML v7.2.6 (Stamatakis 2006) and MrBayes V3.2 (Ronquist & Huelsenbeck 2003), respectively. For BI analysis, the best-fit nucleotide substitution model using the hierarchical likelihood ratio test (hLRT; Huelsenbeck & Crandall 1997; Posada & Crandall 2001) was selected in the MrModelTest 2.2 (Posada & Crandall 1998; Nylander 2004), and the best-fit models for ITS data and the concatenated dataset were (GTR+I+G). In BI analyses, four Markov chains were run from a random starting tree for 3 000 000 generations of ITS dataset and 1 000 000 generations of concatenated dataset, and sampled every 100 generations. The first 25% trees representing the burn-in phase of the analyses were discarded.
2 RESULTS
2.1 Phylogenetic analyses
The combined data set (ITS, nrLSU, and tef1-α) contains 81 sequences originated from 27 specimens representing 14 species, wherein the aligned length of three gene loci is 950, 885, and 580bp, respectively. The topologies are consistent with those of Wang & Zhuang (2019). Gyromitra infula, G. tianshanensis X.C. Wang & W.Y. Zhuang and G. xinjiangensis J.Z. Cao, L. Fan & B. Liu from subgenus Gyromitra grouped together with high support (LB=99%, PP=1.00, Fig. 1), then weakly grouped with G. esculenta and the new species G. venenata. Two collections (HKAS 107322 and HKAS 107322) representing the new species grouped together with high support (LB=100%, PP=1.00), and then clustered with a collection from USA (HMAS 51681) labeled as “G. esculenta” (LB=100%, PP=1.00, Fig. 1).
Fig. 1
Phylogenetic tree inferred from maximum likelihood (ML) analysis based on the combined dataset (ITS, nrLSU, and tef1-α). Only maximum likelihood Bootstraps (LB) over 50% and Bayesian posterior probabilities (PP) over 0.95 are indicated at nodes. Boldfaces exhibit the new species Gyromitra venenata and the epitype for G. esculenta.
The ITS data set includes 74 sequences representing ca. 22 species with an alignment of 960bp. Species from subgenus Gyromitra formed two lineages. Gyromitra infula, G. tianshanensis, and G. xinjiangensis grouped together with high support by both ML and BI analyses (LB=98%, PP=1.00, Fig. 2). Another clade was moderately supported by ML analysis (LB=65%, Fig. 2) and strongly supported by BI analysis (PP=0.96). Three sequences of G. venenata clustered together with moderate support by ML analysis and high support by BI analysis (LB=63%, PP=0.96, Fig. 2) and then grouped with other six sequences recorded as “G. esculenta” from South Korea, USA, Finland and Mexico (LB=86%, PP=0.97, Fig. 2). Gyromitra esculenta, G. splendida and G. tasmanica Berk. & Cooke formed a clade with moderate support by ML analysis and high support by BI analysis (LB=76%, PP=0.99, Fig. 2).
Fig. 2
Phylogenetic tree inferred from ML analysis based on ITS sequences. Only LB over 50% and PP over 0.95 are indicated. Boldfaces exhibit the new species Gyromitra venenata and the epitype for G. esculenta.
2.2 Taxonomy
鹿花菌
Gyromitra esculenta (Pers.) Fr., Summa vegetabilium Scandinaviae 2: 346 (1849). Figs. 3 and 4A, 4B
= Helvella esculenta Pers., Comm. Schaeff. icon. pict.: 64 (1800).
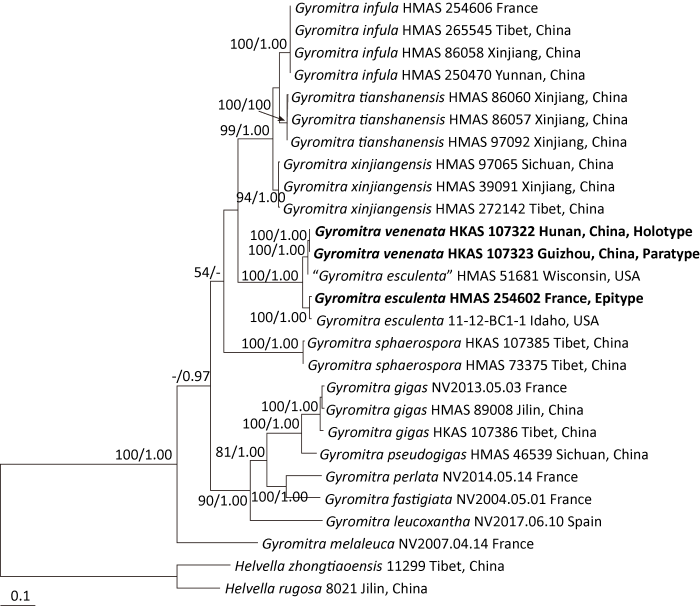
Fig. 3
Microscopic structures of Gyromitra esculenta (drawn from the epitype, HMAS 254602).
A: Ascospores; B: Vertical section of the outmost part of excipulum showing terminal cells on its surface (lower side).
Epitype: FRANCE, 1. Apr 2015, HMAS 214602, here designated.
Gene sequences ex-epitype: MG846990 (ITS), MG847002 (nrLSU), and MG847048 (tef1-α), all of which were generated by Wang & Zhuang (2019).
Excipulum textura intricata, 650-1 000μm thick, composed of 5-10μm wide hyphal elements. Ectal excipulum not well differentiated, outer surface of excipulum sometimes with narrowly clavate to subcylindrical terminal cells, 17-35× 5-7μm (Fig. 3B). Ascospores (14.5-)19-25(-27)× 10.5-13(-13.5)μm [40/2/2], median 22.1×11.82μm, Q=(1.2-)1.65-2.14(-2.29), Qm=1.87±0.17, ellipsoid, nonapiculate, biguttulate; surface finely verrucose to nearly smooth under light microscopy but with irregular wrinkles under SEM (Figs. 3A, 4A, 4B).
Habitat and distribution: On soil in coniferous forests. Widely distributed in Europe, probably also distributed in North America based on sequence data available (Fig. 2).
Additional specimen examined: BULGARIA, HMAS 39743.
毒鹿花菌
Gyromitra venenata Hai J. Li, Z.H. Chen & Zhu L. Yang, sp. nov. Figs. 4C, 5A, 5B, 6
MycoBank MB835549
Diagnosis: Gyromitra venenata is characterized by its stipitate apothecia, cerebriform pileus with a reddish brown, dark reddish brown, brown to dark brown hymenium, a proportionally longer stipe in comparison to the diameter of the pileus, a well-differentiated ectal excipulum with palisade cells, subfusiform, ellipsoid to elongate ellipsoid, nonapiculate, weakly cyanophilic, finely verrucose, biguttulate ascospores (19-28×9.5-12μm) and occurrence in broad-leaved forests dominated by plants of Fagaceae.
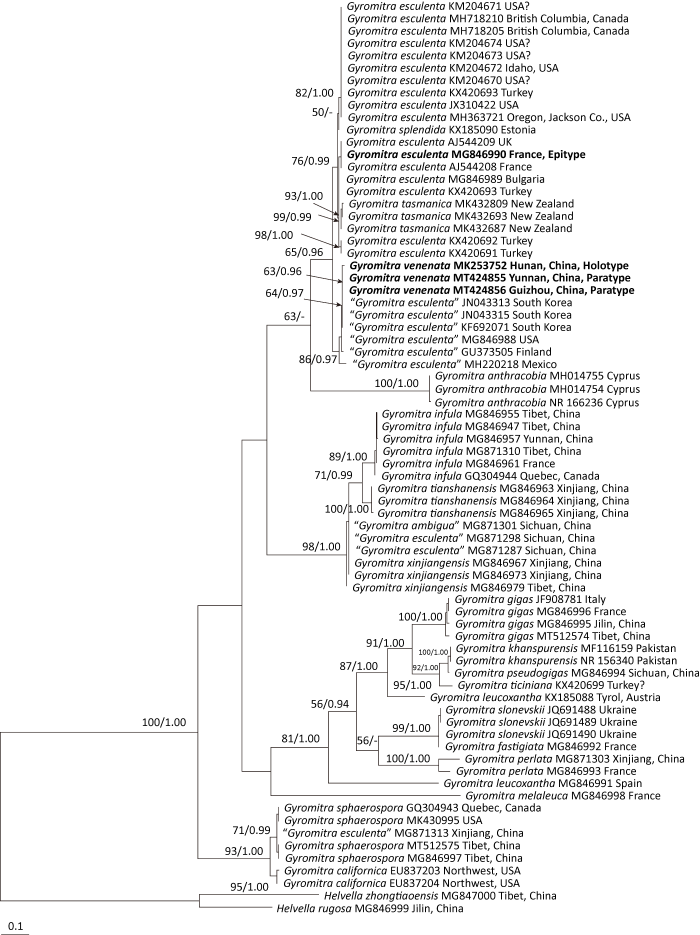
Fig. 4
Scanning electron micrographs of ascospores of Gyromitra esculenta and G. venenata.
A: G. esculenta (HMAS 254602, epitype); B: G. esculenta (HMAS 39743); C: G. venenata (HKAS 107323, paratype).
Holotype: China, Hunan Province, Yanling County, Lingfeng Nature Reserve, 18 Mar 2012, HKAS 107322.
Gene sequences ex-holotype: MK253752 (ITS), MT421931 (nrLSU), and MT424599 (tef1-α).
Etymology: venenata (Lat.) refers to the poisonous ascocarps.
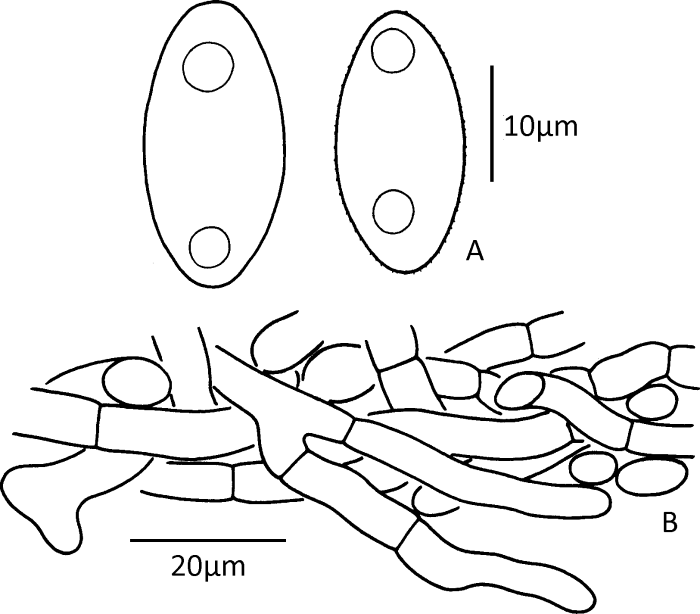
Description: Apothecia stipitate, cerebriform, irregularly lobed (Figs. 5A, 5B). Pileus 1.5-6.7cm high and 2.7-10cm in diam.; margin free or partially connected to surface of stipe; hymenium reddish brown, dark reddish brown, brown to dark brown when fresh, dark brown to almost blackish when dry. Stipe 2.9-6×0.8-3.5cm, cylindrical to subcylindrical, sometimes slightly enlarged at base, white or cream, occasionally pale pinkish or brownish, nearly glabrous, typically fluted with broadly rounded ribs, hollow. Medullary excipulum textura intricata, 400-1 000μm thick, composed of 5-10μm wide hyphal elements. Ectal excipulum textura angularis, 100-250μm thick, composed of ellipsoid to subglobose hyphal elements, 20-40×15-30μm in diam.; terminal cells clavate to subcylindrical, 20-45×7-15μm (Fig. 6C), arranged in a palisade form. Asci subcylindrical to narrowly clavate, 8-spored, uniseriate at maturity, 220-300× 15-19μm (Fig. 6A). Paraphyses erect, frequently septate, moderately branched, subcylindrical; terminal cells subcylindrical or sometimes capitate, 5-8μm in diam., with purplish brown vacuolar and encrusted pigment (Fig. 6A). Ascospores (18-)19-25(-26)×(8-)9.5-12(-13)μm [60/3/3], median 21.57×10.54μm, Q=(1.5-)1.74-2.3(-2.63), Qm=2.06±0.19, ellipsoid to elongate, nonapiculate, weakly cyanophilic, colorless, usually biguttulate, slightly thick-walled (≤0.5μm thick); surface finely verrucose to nearly smooth under light microscopy but with irregular wrinkles under SEM (Figs. 4C, 6B).
Habitat and distribution: Scattered on soil in broad-leaved forests dominated by fagaceous plants. Known from Guizhou, Hunan, and Yunnan provinces, China. Probably widely distributed in the Northern Hemisphere (Fig. 2).
Additional specimens examined: CHINA, Guizhou Province, Zunyi, Xishui County, 13 Mar 2020, HKAS 107323, MHHNU 31587 and NIOPH GZ20200313-01. Hunan Province, Zhuzhou, Yanling County, Lingfeng Nature Reserve, 18 Mar 2012, MHHNU 7671 (Isotype, regarded as G. esculenta by Chen et al. 2016; Chen & Zhang 2019). Yunnan Province, Zhaotong, Zhenxiong County, Luokan Town, Damiao Village, Huocaoping Group, Matangwan, 27°45.4′N, 104°51.8′E, 1 200- 1 300m alt., 26 Mar 2020, HKAS 107324, NIOPH 2X-001.
Fig. 5
Apothecia of poisonous or potentially poisonous species of Gyromitra.
A, B: Gyromitra venenata (holotype); C, D: Gyromitra gigas (HKAS 107386); E, F: Gyromitra sphaerospora (HKAS 107385). Bars=2cm.
Fig. 6
Microscopic structures of Gyromitra venenata (drawn from the holotype).
A: Upper parts of paraphyses and ascus; B: Ascospores; C: Vertical section of the outmost part of the ectal excipulum showing terminal cells arranged in a palisade form.
3 DISCUSSION
Gyromitra venenata is very similar to G. esculenta morphologically and a sister taxon of the latter phylogenetically (Figs. 1 and 2). However, the new species has a well differentiated ectal excipulum with palisade cells, a proportionally longer stipe and a distribution in broad-leaved forests. On the contrary, G. esculenta has a one-layered excipulum, a proportionally shorter stipe and a distribution in coniferous forests (Hansen & Knudsen 2000; van Vooren & Moreau 2009b; Perić & Perić 2010; Methven et al. 2013). The interspecific variation of ITS sequence between G. esculenta and G. venenata is 8.22% (65/791bp). Our phylogenetic analysis indicated that G. venenata, and G. gigas (Fig. 2; Fig. 5A-5D), should be widely distributed in the Northern Hemisphere, a phenomenon common in Pezizales (Du et al. 2012; Wang & Zhuang 2019).
Gyromitra venenata is also somewhat similar to G. infula, G. tianshanensis and G. xinjiangensis. However, the pileus of G. infula is saddle-shaped with two prominent lobes and its ascospores is distinctly narrower (19-24×7-8.5μm, van Vooren & Moreau 2009b; Methven et al. 2013). Gyromitra tianshanensis, a new species descried from China, also has a brain-like pileus. However, the former species has smaller ascospores (17-21.5×6-9.5μm) and mainly occurs on rotten wood in summer (Wang & Zhuang 2019). Gyromitra xinjiangensis differs from G. venenata by its paraphyses with hamate apex, verrucose ascospores (20-25× 8-11μm), distributed in coniferous forests (mainly Picea) at higher altitudes (2 500-3 300m) in summer to autumn (Cao et al. 1990; Wang & Zhuang 2019).
Gyromitra ambigua (P. Karst.) Harmaja is somewhat similar to G. venenata by its smooth to finely verrucose, biguttulate ascospores, but the former species has a saddle-shaped pileus, fusiform to oblong ellipsoid ascospores with obviously rounded apiculus at each end (Cao et al. 1990; Carlsen & Stensrud 2003; van Vooren & Moreau 2009b).
Gyromitra lactea J.Z. Cao, L. Fan & B. Liu, G. pseudogigas X.C. Wang & W.Y. Zhuang and G. sichuanensis Korf & W.Y. Zhuang were originally described from China (Korf & Zhuang 1985; Cao et al. 1990; Wang & Zhuang 2019), and should be compared with the new species. However, G. lactea differs from G. venenata by its cream, discoid apothecia, mostly uniguttulate, rarely biguttulate ascospores and grows on rotten wood in summer (Cao et al. 1990). Gyromitra pseudogigas is easily distinguished from G. venenata by its saddle-shaped apothecia, finely roughened, uniguttulate or triguttulate apothecia with prominent apiculus at each end, and the type specimen was collected in July (Wang & Zhuang 2019). Gyromitra sichuanensis differs from G. venenata by its paraphyses with hamate apex, verrucose ascospores and distributed in coniferous forests (Korf & Zhuang 1985; Wang & Zhuang 2019).
Gyromitra discinoides (S. Imai) S. Imai, originally described from Hokkaido, Japan, differs from G. venenata by its cinnamon to dark brown pileus, deeply fluted stipe, wider ascospores (21-28×11-15μm, Imai 1932, 1954). Gyromitra longipes Harmaja is similar to G. venenata by its cerebriform pileus and proportionally longer stipe in comparison to the diameter of the pileus, but the former species has wider asci (210-290× 20-28μm), larger ascospores (22-32×10-14μm) with broadly rounded apiculus at each end and is distributed in North Europe (Harmaja 1979; Carlsen & Stensrud 2003; van Vooren & Moreau 2009b). Gyromitra brunnea Underw., originally described from North America, is more or less similar to G. venenata by its pinkish brown, reddish brown or tan color pileus, but the former species usually has a saddle-shaped pileus, and distinctly warty ascospores (22-29×10-13.5μm) with ornaments 1-2μm long from the surface of the spore or 2-5μm long at the ends of the spore, where projections appear as 1-5 apiculi (Underwood 1893; Kimbrough et al. 1990; Methven et al. 2013; Acar et al. 2018).
Our phylogenetic analysis (Fig. 2) indicated that the phylogeny and taxonomy of G. esculenta-G. tasmanica-G. splendida merit further study in the near future. The occurrence of G. esculenta in China has not been confirmed by molecular evidence yet, although this species was repeatedly reported from China in the past (Tai 1979; Mao 2006; Bau et al. 2014; Chen et al. 2014, 2016; Li et al. 2015; Chen & Zhang 2019; Wu et al. 2019). Cao & Zhu (1992) didn’t find any typical collections of G. esculenta from China. Ekanayaka et al. (2018) reported “G. esculenta” from Xinjiang, China, based on a collection, namely HKAS 87684. Our phylogenetic analysis and morphological reexamination of the collection indicated that it is G. sphaerospora (Peck) Sacc. (Fig. 2; Fig. 5E, 5F). Other two ITS sequences (MG871287 and MG871298) submitted to GenBank labeled as “G. esculenta” by the same authors were revealed to be G. xinjiangensis by the present study (Fig. 2). The ITS sequence MG871287 was assigned to HKAS 85347 (Ekanayaka et al. 2018). However, we found that HKAS 85347 is actually Tuber sinoaestivum J.P. Zhang & P.G. Liu. Another specimen (HKAS 82861) was erroneously cited as “HK036” and regarded as G. ambigua (Ekanayaka et al. 2018) is in fact G. xinjiangensis (Fig. 2).
Gyromitra esculenta has caused many severe poisoning incidents and even deaths (Michelot & Toth 1991; Leathem & Dorran 2007; Patocka et al. 2012; Arłukowicz-Grabowska et al. 2019). It is wise to avoid collecting and eating this species, and even its sale or trading is considered to be illegal in some European countries (Ludolph 2000). To the best of our knowledge, G. venenata has caused two poisoning incidents in China, with syndromes similar to those produced by G. esculenta. Therefore, we strongly recommend the public do not eat this species.
参考文献
The Hevellaceae: systematic revision and occurrence in northern and northwestern North America
Morphology and phylogeny reveal a new record Gyromitra for Turkish mycobiota
Acute liver injury, acute liver failure and acute on chronic liver failure: a clinical spectrum of poisoning due to Gyromitra esculenta
DOI:10.1016/j.aohep.2018.11.009
URL
PMID:31014949
[本文引用: 3]

Gyromitra esculenta, also known as
A revised checklist of poisonous mushrooms in China
Hattmorkelen Gyromitra longipes Harmaja funnet i Norge
Notes on the genus Gyromitra from China
Gyromitra esculenta and its identification
Investigation and analysis of 102 mushroom poisoning cases in Southern China from 1994 to 2012
Multiple sequence alignment with the Clustal series of programs
DOI:10.1093/nar/gkg500
URL
PMID:12824352
[本文引用: 1]

The Clustal series of programs are widely used in molecular biology for the multiple alignment of both nucleic acid and protein sequences and for preparing phylogenetic trees. The popularity of the programs depends on a number of factors, including not only the accuracy of the results, but also the robustness, portability and user-friendliness of the programs. New features include NEXUS and FASTA format output, printing range numbers and faster tree calculation. Although, Clustal was originally developed to run on a local computer, numerous Web servers have been set up, notably at the EBI (European Bioinformatics Institute) (http://www.ebi.ac.uk/clustalw/).
Fungal Planet description sheets: 716-784
DOI:10.3767/persoonia.2018.40.10
URL
PMID:30505003
[本文引用: 3]

Novel species of fungi described in this study include those from various countries as follows: Australia, Chaetopsina eucalypti on Eucalyptus leaf litter, Colletotrichum cobbittiense from Cordyline stricta x C. australis hybrid, Cyanodermella banksiae on Banksia ericifolia subsp. macrantha, Discosia macrozamiae on Macrozamia miquelii, Elsinoe banksiigena on Banksia marginata, Elsinoe elaeocarpi on Elaeocarpus sp., Elsinoe leucopogonis on Leucopogon sp., Helminthosporium livistonae on Livistona australis, Idriellomyces eucalypti (incl. Idriellomyces gen. nov.) on Eucalyptus obliqua, Lareunionomyces eucalypti on Eucalyptus sp., Myrotheciomyces corymbiae (incl. Myrotheciomyces gen. nov., Myrotheciomycetaceae fam. nov.), Neolauriomyces eucalypti (incl. Neolauriomyces gen. nov., Neolauriomycetaceae fam. nov.) on Eucalyptus sp., Nullicamyces eucalypti (incl. Nullicamyces gen. nov.) on Eucalyptus leaf litter, Oidiodendron eucalypti on Eucalyptus maidenii, Paracladophialophora cyperacearum (incl. Paracladophialophoraceae fam. nov.) and Periconia cyperacearum on leaves of Cyperaceae, Porodiplodia livistonae (incl. Porodiplodia gen. nov., Porodiplodiaceae fam. nov.) on Livistona australis, Sporidesmium melaleucae (incl. Sporidesmiales ord. nov.) on Melaleuca sp., Teratosphaeria sieberi on Eucalyptus sieberi, Thecaphora australiensis in capsules of a variant of Oxalis exilis. Brazil, Aspergillus serratalhadensis from soil, Diaporthe pseudoinconspicua from Poincianella pyramidalis, Fomitiporella pertenuis on dead wood, Geastrum magnosporum on soil, Marquesius aquaticus (incl. Marquesius gen. nov.) from submerged decaying twig and leaves of unidentified plant, Mastigosporella pigmentata from leaves of Qualea parviflorae, Mucor souzae from soil, Mycocalia aquaphila on decaying wood from tidal detritus, Preussia citrullina as endophyte from leaves of Citrullus lanatus, Queiroziella brasiliensis (incl. Queiroziella gen. nov.) as epiphytic yeast on leaves of Portea leptantha, Quixadomyces cearensis (incl. Quixadomyces gen. nov.) on decaying bark, Xylophallus clavatus on rotten wood. Canada, Didymella cari on Carum carvi and Coriandrum sativum. Chile, Araucasphaeria foliorum (incl. Araucasphaeria gen. nov.) on Araucaria araucana, Aspergillus tumidus from soil, Lomentospora valparaisensis from soil. Colombia, Corynespora pseudocassiicola on Byrsonima sp., Eucalyptostroma eucalyptorum on Eucalyptus pellita, Neometulocladosporiella eucalypti (incl. Neometulocladosporiella gen. nov.) on Eucalyptus grandis x urophylla, Tracylla eucalypti (incl. Tracyllaceae fam. nov., Tracyllalales ord. nov.) on Eucalyptus urophylla. Cyprus, Gyromitra anthracobia (incl. Gyromitra subg. Pseudoverpa) on burned soil. Czech Republic, Lecanicillium restrictum from the surface of the wooden barrel, Lecanicillium testudineum from scales of Trachemys scripta elegans. Ecuador, Entoloma yanacolor and Saproamanita quitensis on soil. France, Lentithecium carbonneanum from submerged decorticated Populus branch. Hungary, Pleuromyces hungaricus (incl. Pleuromyces gen. nov.) from a large Fagus sylvatica log. Iran, Zymoseptoria crescenta on Aegilops triuncialis. Malaysia, Ochroconis musicola on Musa sp. Mexico, Cladosporium michoacanense from soil. New Zealand , Acrodontium metrosideri on Metrosideros excelsa, Polynema podocarpi on Podocarpus totara, Pseudoarthrographis phlogis (incl. Pseudoarthrographis gen. nov.) on Phlox subulata. Nigeria, Coprinopsis afrocinerea on soil. Pakistan, Russula mansehraensis on soil under Pinus roxburghii. Russia, Baorangia alexandri on soil in deciduous forests with Quercus mongolica. South Africa, Didymocyrtis brachylaenae on Brachylaena discolor. Spain, Alfaria dactylis from fruit of Phoenix dactylifera, Dothiora infuscans from a blackened wall, Exophiala nidicola from the nest of an unidentified bird, Matsushimaea monilioides from soil, Terfezia morenoi on soil. United Arab Emirates, Tirmania honrubiae on soil. USA, Arxotrichum wyomingense (incl. Arxotrichum gen. nov.) from soil, Hongkongmyces snookiorum from submerged detritus from a fresh water fen, Leratiomyces tesquorum from soil, Talaromyces tabacinus on leaves of Nicotiana tabacum. Vietnam, Afroboletus vietnamensis on soil in an evergreen tropical forest, Colletotrichum condaoense from Ipomoea pes-caprae. Morphological and culture characteristics along with DNA barcodes are provided.
Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China
DOI:10.1016/j.fgb.2012.03.006
URL
[本文引用: 1]

The phylogenetic diversity of true morels (Morchella) in China was estimated by initially analyzing nuclear ribosomal internal transcribed spacer (ITS) rDNA sequences from 361 specimens collected in 21 provinces during the 2003-2011 growing seasons, together with six collections obtained on loan from three Chinese herbaria. Based on the results of this preliminary screen, 40 Esculenta Clade (yellow morels) and 30 Elata Clade (black morels) were chosen to represent the full range of phylogenetic diversity sampled. To investigate their species limits, we generated DNA sequences from portions of three protein-coding genes (RPB1, RPB2 and EF-1 alpha) and domains D1 and D2 of the nuclear large subunit (LSU) rDNA for all 70 collections. To fully assess evolutionary relationships, previously published multilocus DNA sequence data representing all known Morchella species was included in this study. Phylogenetic analyses employing maximum parsimony and maximum likelihood frameworks resolved 30 species in China compared with 22 in Europe and 19 within North America. Eleven novel phylogenetically distinct species were discovered in China, including two species within the Elata Clade and nine within the Esculenta Clade. Of the 30 species in China, 20 appear to be endemic, nine were also represented in Europe, and four putatively fire-adapted species have disjunct distributions in China, Europe and western North America. Although the diversification time estimates place the Esculenta Clade in China as early as the late Cretaceous and the Elata Clade by the early Oligocene, 27 of the 30 species evolved between the middle Miocene 12 Mya and present. (C) 2012 Elsevier Inc.
Biotrophic associations between lodgepole pine seedlings and postfire ascomycetes (Pezizales) in monoxenic culture
DOI:10.1139/b86-359 URL [本文引用: 1]
Taxonomy and phylogeny of operculate discomycetes: Pezizomycetes
DOI:10.1007/s13225-018-0402-z URL [本文引用: 3]
ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts
A case of fatal poisoning by Gyromitra esculenta
DOI:10.1007/BF00297052 URL PMID:4480349 [本文引用: 1]
BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT
Systematics of the Pezizomycetes-the operculate discomycetes
DOI:10.3852/mycologia.98.6.1029
URL
PMID:17486978
[本文引用: 1]

The Pezizomycetes (order Pezizales) is an early diverging lineage within the Pezizomycotina. A shared derived character, the operculate ascus, supports the Pezizales as monophyletic, although functional opercula have been lost in certain taxa. Phylogenetic relationships within Pezizales were studied using parsimony and Bayesian analyses of partial SSU and LSU rDNA sequences from 100 taxa representing 82 genera and 13 of the 15 families currently recognized. Three primary lineages are identified that more or less correspond to the A, B and C lineages resolved in previous analyses using SSU rDNA: (A) Ascobolaceae and Pezizaceae; (B) Discinaceae-Morchellaceae and Helvellaceae-Tuberaceae; (C) Ascodesmidaceae, Glaziellaceae, Pyronemataceae, Sarcoscyphaceae and Sarcosomataceae. In contrast the monotypic Rhizinaceae and Caloscyphaceae are resolved as two independent lineages. Bayesian analyses support a relationship among Rhizina and two species of Psilopezia (Pyronemataceae). Only lineage C is highly supported. The B and C lineages form a strongly supported monophyletic group. None of these lineages corresponds to earlier proposed suborders. The A and B lineages are supported by certain morphological features (e.g. ascus bluing reaction in iodine, cytology of spores and paraphyses, septal pore structures and excipulum structure); these characters have been subject to homoplasy. Lineage C is the largest and most heterogeneous, and no unifying morphological features support its recognition. The Pyronemataceae, in which almost half of the species in the order are found, is not monophyletic because the Ascodesmidaceae and Glaziellaceae are nested within it. The relationships among all families in the C lineage remain uncertain. The origin of various forms of ascomata, including hypogeous forms (truffles and truffle-like), epigeous cleistothecia, simple reduced apothecia and highly elaborate, stipitate forms (helvelloid and morchelloid), are discussed.
Nordic macromycetes. Vol. 1 Ascomycetes
Amendments of the limits of the genera Gyromitra and Pseudorhizina, with the description of a new species, Gyromitra montana
Studies on vernal species of Gyromitra and Pseudombrophila (syn. Nannfeldtiella)
Phylogeny estimation and hypothesis testing using maximum likelihood
Contribution to the knowledge of the classification of Helvellaceae
Elvellaceae Japoniae
Ultrastructural observations on Helvellaceae (Pezizales, Ascomycetes). IV. Ascospore ontogeny in selected species of Gyromitra subgenus Discina
Dictionary of the fungi. 10th ed
Some new species and new records of discomycetes in China
Fungal systematics and evolution: FUSE 3
DOI:10.12905/0380.sydowia69-2017-0229
URL
PMID:29386695
[本文引用: 1]

The present study introduces seven new species, one new combination, one new variety and several interesting taxonomical notes and/or geographical records. Most of the new taxa are Ascomycetes, but the study also includes a new variety of a Basidiomycete. Novel species include Gyromitra khanspurensis (Discinaceae, Pezizales, Pezizomycetes) from Pakistan growing near Cedrus deoadara and Paramyrothecium guiyangense and Paramyrothecium verruridum (Stachybotriaceae, Hypocreales, Sordariomycetes) both isolated from soil in China. New species from South Africa are Sclerostagonospora elegiae on culm litter of Elegia equisetacea, Sclerostagonospora fusiformis on culm litter of Thamnochortus spicigerus, Sclerostagonospora pinguis on culm litter of Cannomois virgata and Sclerostagonospora sulcata on culm litter of Ischyrolepis subverticellata (Phaeosphaeriaceae, Pleosporales, Dothideomycetes). Hapalocystis berkeleyi var. kickxii with its basionym Hypoxylon kickxii is shown to be a taxon on species level and thus recombined as Hapalocystis kickxii (Sydowiellaceae, Diaporthales, Sordariomycetes), and it is lecto- and epitypified. The new variety Pluteus romellii var. luteoalbus (Pluteaceae, Agaricales, Agaricomycetes) growing on a mossy fallen stem of a deciduous tree is described from Czech Republic. Cortinarius scaurocaninus (Cortinariaceae, Agaricales, Agaricomycetes) is new for Austria, Humicola grisea (Chaetomiaceae, Sordariales, Sordariomycetes) is an interesting new record for Chile. Two taxa are reported as new for Turkey: the lichenicolous fungus Opegrapha parasitica (Opegraphaceae, Arthoniales, Arthoniomycetes) growing partly immersed in the thallus of Aspicilia and the lichen Rinodina zwackhiana (Physciaceae, Teloschistales, Lecanoromycetes) from calcareous rock. Finally, Xerula strigosa (Physalacriaceae, Agaricales, Agaricomycetes), described from China, is confirmed to be present also in Pakistan.
Poisoning due to raw Gyromitra esculenta (false morels) west of the Rockies
DOI:10.1017/s1481803500014937
URL
PMID:17391587
[本文引用: 3]

Vomiting with abdominal pain is a common presentation in the emergency department (ED). Without a careful history, unusual causes, such as toxic ingestion, may evade diagnosis. We report a case of an Asian couple who presented to the ED with vomiting and epigastric distress. They were discharged with no definite diagnosis, but on a return ED visit the following day were diagnosed with toxic ingestion of Gyromitra esculenta, commonly known as the western false morel. The patients were admitted and treated with intravenous hydration and pyridoxine. Both patients developed mild hepatotoxicity but went on to fully recover. This case demonstrates that the western false morel may cause significant toxicity and it highlights the importance of obtaining a complete history in patients who present with non-specific gastrointestinal symptoms.
Poisonous mushrooms and their toxins in China
A molecular phylogenetic assessment of the genus Gyromitra in North America
DOI:10.3852/12-397
URL
[本文引用: 8]

Gyromitra is a widespread genus of macroscopic apothecial ascomycetes whose taxa may be mycorrhizal, saprophytic or parasitic. Nuclear ribosomal 28S large subunit sequence data from 35 specimens from North America, along with sequences available in GenBank, were used in maximum-parsimony, maximum-likelihood and Bayesian analyses to reconstruct a phylogeny of Gyromitra in North America. Gyromitra sensu lato forms a monophyletic group within the Discinaceae composed of five distinct subgenera and 11 well supported clades that include Discina, Hydnotrya and Pseudorhizina. A new subgenus is proposed to accommodate G. californica and G. sphaerospora.
Poisoning by Gyromitra esculenta-a review
DOI:10.1002/jat.2550110403
URL
PMID:1939997
[本文引用: 3]

Gyromitra esculenta (Pers.: Fr.) Fr. and a few other mushrooms have caused severe poisonings and even deaths in humans. Clinical data are characterized primarily by vomiting and diarrhoea, followed by jaundice, convulsions and coma. Gastrointestinal disorders distinguish this poisoning. Frequent consumption can cause hepatitis and neurological diseases. The species of concern are mainly G. esculenta and G. gigas (Kromb.) Cooke (non Phill.). Nevertheless, recent advances in chromatography, biochemistry and toxicology have established that other Ascomycetes species also may prove toxic. Gyromitrin (acetaldehyde methylformylhydrazone, G) and its homologues are toxic compounds that convert in vivo into N-methyl-N-formylhydrazine (MFH), and then into N-methylhydrazine (MH). The toxicity of these chemicals, which are chiefly hepatotoxic and even carcinogenic, has been established through in vivo and in vitro experiments using animals, cell cultures and biochemical systems. When we consider the chemical nature and the reactivity of these natural compounds, we suggest that chemical and biochemical mechanisms may explain their intrinsic biological activity.
MrModeltest 2.2. Program distributed by the author
Gyromitrin, mushroom toxin of Gyromitra spp
DOI:10.31482/mmsl.2012.008 URL [本文引用: 3]
Gyromitra Fr. sensu lato (Discinaceae, Pezizales) in Montenegro
Modeltest: testing the model of DNA substitution
DOI:10.1093/bioinformatics/14.9.817
URL
PMID:9918953
[本文引用: 1]

SUMMARY: The program MODELTEST uses log likelihood scores to establish the model of DNA evolution that best fits the data. AVAILABILITY: The MODELTEST package, including the source code and some documentation is available at http://bioag.byu. edu/zoology/crandall_lab/modeltest.html.
Selecting the best-fit model of nucleotide substitution
Despite the relevant role of models of nucleotide substitution in phylogenetics, choosing among different models remains a problem. Several statistical methods for selecting the model that best fits the data at hand have been proposed, but their absolute and relative performance has not yet been characterized. In this study, we compare under various conditions the performance of different hierarchical and dynamic likelihood ratio tests, and of Akaike and Bayesian information methods, for selecting best-fit models of nucleotide substitution. We specifically examine the role of the topology used to estimate the likelihood of the different models and the importance of the order in which hypotheses are tested. We do this by simulating DNA sequences under a known model of nucleotide substitution and recording how often this true model is recovered by the different methods. Our results suggest that model selection is reasonably accurate and indicate that some likelihood ratio test methods perform overall better than the Akaike or Bayesian information criteria. The tree used to estimate the likelihood scores does not influence model selection unless it is a randomly chosen tree. The order in which hypotheses are tested, and the complexity of the initial model in the sequence of tests, influence model selection in some cases. Model fitting in phylogenetics has been suggested for many years, yet many authors still arbitrarily choose their models, often using the default models implemented in standard computer programs for phylogenetic estimation. We show here that a best-fit model can be readily identified. Consequently, given the relevance of models, model fitting should be routine in any phylogenetic analysis that uses models of evolution.
A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs
DOI:10.3852/mycologia.97.1.84
URL
PMID:16389960
[本文引用: 1]

Beauveria is a globally distributed genus of soil-borne entomopathogenic hyphomycetes of interest as a model system for the study of entomopathogenesis and the biological control of pest insects. Species recognition in Beauveria is difficult due to a lack of taxonomically informative morphology. This has impeded assessment of species diversity in this genus and investigation of their natural history. A gene-genealogical approach was used to investigate molecular phylogenetic diversity of Beauveria and several presumptively related Cordyceps species. Analyses were based on nuclear ribosomal internal transcribed spacer (ITS) and elongation factor 1-alpha (EF1-alpha) sequences for 86 exemplar isolates from diverse geographic origins, habitats and insect hosts. Phylogenetic trees were inferred using maximum parsimony and Bayesian likelihood methods. Six well supported clades within Beauveria, provisionally designated A-F, were resolved in the EF1-alpha and combined gene phylogenies. Beauveria bassiana, a ubiquitous species that is characterized morphologically by globose to subglobose conidia, was determined to be non-monophyletic and consists of two unrelated lineages, clades A and C. Clade A is globally distributed and includes the Asian teleomorph Cordyceps staphylinidaecola and its probable synonym C. bassiana. All isolates contained in Clade C are anamorphic and originate from Europe and North America. Clade B includes isolates of B. brongniartii, a Eurasian species complex characterized by ellipsoidal conidia. Clade D includes B. caledonica and B. vermiconia, which produce cylindrical and comma-shaped conidia, respectively. Clade E, from Asia, includes Beauveria anamorphs and a Cordyceps teleomorph that both produce ellipsoidal conidia. Clade F, the basal branch in the Beauveria phylogeny includes the South American species B. amorpha, which produces cylindrical conidia. Lineage diversity detected within clades A, B and C suggests that prevailing morphological species concepts underestimate species diversity within these groups. Continental endemism of lineages in B. bassiana s.l. (clades A and C) indicates that isolation by distance has been an important factor in the evolutionary diversification of these clades. Permutation tests indicate that host association is essentially random in both B. bassiana s.l. clades A and C, supporting past assumptions that this species is not host specific. In contrast, isolates in clades B and D occurred primarily on coleopteran hosts, although sampling in these clades was insufficient to assess host affliation at lower taxonomic ranks. The phylogenetic placement of Cordyceps staphylinidaecola/bassiana, and C. scarabaeicola within Beauveria corroborates prior reports of these anamorph-teleomorph connections. These results establish a phylogenetic framework for further taxonomic, phylogenetic and comparative biological investigations of Beauveria and their corresponding Cordyceps teleomorphs.
MrBayes 3: Bayesian phylogenetic inference under mixed models
DOI:10.1093/bioinformatics/btg180
URL
PMID:12912839
[本文引用: 1]

MrBayes 3 performs Bayesian phylogenetic analysis combining information from different data partitions or subsets evolving under different stochastic evolutionary models. This allows the user to analyze heterogeneous data sets consisting of different data types-e.g. morphological, nucleotide, and protein-and to explore a wide variety of structured models mixing partition-unique and shared parameters. The program employs MPI to parallelize Metropolis coupling on Macintosh or UNIX clusters.
RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models
DOI:10.1093/bioinformatics/btl446
URL
PMID:16928733
[本文引用: 1]

UNLABELLED: RAxML-VI-HPC (randomized axelerated maximum likelihood for high performance computing) is a sequential and parallel program for inference of large phylogenies with maximum likelihood (ML). Low-level technical optimizations, a modification of the search algorithm, and the use of the GTR+CAT approximation as replacement for GTR+Gamma yield a program that is between 2.7 and 52 times faster than the previous version of RAxML. A large-scale performance comparison with GARLI, PHYML, IQPNNI and MrBayes on real data containing 1000 up to 6722 taxa shows that RAxML requires at least 5.6 times less main memory and yields better trees in similar times than the best competing program (GARLI) on datasets up to 2500 taxa. On datasets > or =4000 taxa it also runs 2-3 times faster than GARLI. RAxML has been parallelized with MPI to conduct parallel multiple bootstraps and inferences on distinct starting trees. The program has been used to compute ML trees on two of the largest alignments to date containing 25,057 (1463 bp) and 2182 (51,089 bp) taxa, respectively. AVAILABILITY: icwww.epfl.ch/~stamatak
List of cryptogams at present known to inhabit the State of Indiana
Essai taxinomique sur le genre Gyromitra Fr. sensu lato (Pezizales). 1. Introduction et systématique
Essai taxinomique sur le genre Gyromitra Fr. sensu lato (Pezizales). 2. Le genre Gyromitra Fr., sous-genre Gyromitra
Essai taxinomique sur le genre Gyromitra Fr. sensu lato (Pezizales). 3. Le genre Gyromitra Fr., sous-genre Discina
Essai taxinomique sur le genre Gyromitra Fr. sensu lato (Pezizales). 4. Le genre Gyromitra Fr., sous-genre Caroliniana
Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species
DOI:10.1128/jb.172.8.4238-4246.1990
URL
PMID:2376561
[本文引用: 1]

Detailed restriction analyses of many samples often require substantial amounts of time and effort for DNA extraction, restriction digests, Southern blotting, and hybridization. We describe a novel approach that uses the polymerase chain reaction (PCR) for rapid simplified restriction typing and mapping of DNA from many different isolates. DNA fragments up to 2 kilobase pairs in length were efficiently amplified from crude DNA samples of several pathogenic Cryptococcus species, including C. neoformans, C. albidus, C. laurentii, and C. uniguttulatus. Digestion and electrophoresis of the PCR products by using frequent-cutting restriction enzymes produced complex restriction phenotypes (fingerprints) that were often unique for each strain or species. We used the PCR to amplify and analyze restriction pattern variation within three major portions of the ribosomal DNA (rDNA) repeats from these fungi. Detailed mapping of many restriction sites within the rDNA locus was determined by fingerprint analysis of progressively larger PCR fragments sharing a common primer site at one end. As judged by PCR fingerprints, the rDNA of 19 C. neoformans isolates showed no variation for four restriction enzymes that we surveyed. Other Cryptococcus spp. showed varying levels of restriction pattern variation within their rDNAs and were shown to be genetically distinct from C. neoformans. The PCR primers used in this study have also been successfully applied for amplification of rDNAs from other pathogenic and nonpathogenic fungi, including Candida spp., and ought to have wide applicability for clinical detection and other studies.
A three-locus phylogeny of Gyromitra (Discinaceae, Pezizales) and discovery of two cryptic species
DOI:10.1080/00275514.2018.1515456
URL
PMID:30543486
[本文引用: 11]

Gyromitra species are known as
Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species
DOI:10.1007/s13225-019-00432-7 URL [本文引用: 7]
Molecular phylogenetic analyses redefine seven major clades and reveal 22 new generic clades in the fungal family Boletaceae
DOI:10.1007/s13225-014-0283-8
URL
[本文引用: 2]

Mushrooms in the basidiomycete family Boletaceae are ecologically and economically very important. However, due to the morphological complexity and the limited phylogenetic information on the various species and genera of this fungal family, our understanding of its systematics and evolution remains rudimentary. In this study, DNA sequences of four genes (nrLSU, tef1-alpha, rpb1, and rpb2) were newly obtained from ca. 200 representative specimens of Boletaceae. Our phylogenetic analyses revealed seven major clades at the subfamily level, namely Austroboletoideae, Boletoideae, Chalciporoideae, Leccinoideae, Xerocomoideae, Zangioideae, and the Pulveroboletus Group. In addition, 59 genus-level clades were identified, of which 22 were uncovered for the first time. These 22 clades were mainly placed in Boletoideae and the Pulveroboletus Group. The results further indicated that the characters frequently used in the morphology-based taxonomy of Boletaceae, such as basidiospore ornamentation, the form of the basidioma, and the stuffed pores each had multiple origins within the family, suggesting that the use of such features for high-level classification of Boletaceae should be de-emphasized and combined with other characters.
One hundred noteworthy boletes from China
DOI:10.1007/s13225-016-0375-8 URL [本文引用: 1]