近年来,由于生活方式和医疗手段的改变,人口老龄化和免疫受损/抑制人群的数量激增,导致真菌感染问题日益严重。每年约有150万人死于侵袭性真菌感染,其中以白念珠菌为首的念珠菌感染数量每年超过40万例,位居全球侵袭性真菌感染前三(Brown et al. 2012)。随着广谱抗生素、抗真菌药物和现代医疗技术(如免疫抑制和器官移植等)的广泛应用,耐药问题也日益严重,但新的病原真菌却在不断出现,其中引起关注度最大的是多重耐药真菌——耳念珠菌的出现。
耳念珠菌Candida auris为近年来发现的新兴致病性念珠菌,其2009年首次在日本患者的耳道中分离到(Satoh et al. 2009),随后由耳念珠菌引起的感染在全球范围迅速蔓延,对全球公共卫生健康造成严重威胁。随之对耳念珠菌的生物学研究也相继展开。与其他念珠菌相比,耳念珠菌容易通过医疗保健设施进行传播,从而造成院内感染的暴发(Schelenz et al. 2016;Chowdhary et al. 2017)。耳念珠菌在人类宿主和无机体表面均能存活很长时间(Abdolrasouli et al. 2017;Welsh et al. 2017);此外,90%的耳念珠菌临床菌株对氟康唑具有耐药性,对其他唑类抗真菌药物、5-氟胞嘧啶(Rhodes et al. 2018)、两性霉素B(Escandon et al. 2019)和棘白菌素类(Berkow & Lockhart 2018;Chowdhary et al. 2018;Rhodes et al. 2018)的敏感性也多变,所以耳念珠菌被普遍认为是多重耐药病原真菌(Chowdhary et al. 2014;Kathuria et al. 2015)。此外,耳念珠菌容易被错误鉴定为其他念珠菌,这为临床用药带来问题,这些都促使耳念珠菌院内感染在全世界范围不断扩大。总之,耳念珠菌具有多重耐药、高致死率、易于院内传播以及鉴定困难等特征,被冠以“超级真菌”的称号。本文将对耳念珠菌在全球的感染现状进行总结,比较耳念珠菌与最常见的念珠菌属病原真菌的差异和相似性,同时结合目前对其生物学的研究进展进行综述,以期帮助人们更好地认识这一新兴病原真菌。
1 耳念珠菌感染的流行病学
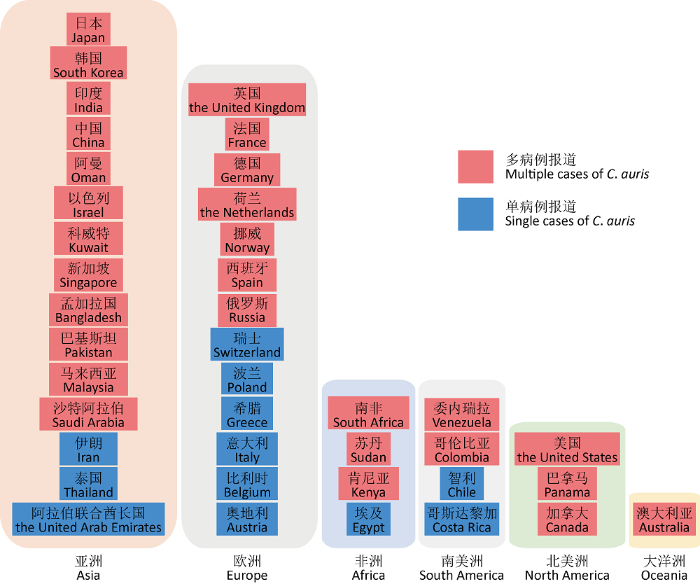
2009年,日本科学家首次报道从一名70岁女性患者的外耳道中分离到一株子囊菌,通过形态学、代谢特征以及系统发育分析后发现,其为一新兴病原真菌,他们将其命名为耳念珠菌(Satoh et al. 2009)。随后,全球不同地域先后都从不同的样本中分离到耳念珠菌。根据美国疾控中心的数据统计和文献检索,截止2020年7月,全球除南极洲外的6大洲,共计40个国家报道了耳念珠菌感染或携带病例,其中12个国家为单病例报道,28个国家为多病例报道(https://www.cdc.gov/,图1)。
图1
图1
截止2020年7月全球报道耳念珠菌感染或携带病例的国家
蓝色背景标注为单病例报道国家,红色背景标注为多病例报道国家
Fig. 1
Countries with reported cases of Candida auris infection or carriage from 2009 to July 2020.
The countries with single case were highlighted in blue and those with multiple cases were highlighted in red.
1.1 亚洲
目前,亚洲多达15个国家出现了耳念珠菌感染病例。2009年,除了日本首次报道耳念珠菌外,韩国也从中耳炎患者耳道中分离到15株耳念珠菌(Kim et al. 2009);2011年韩国又报道了3例由耳念珠菌引起的院内真菌败血症,其中一例为回顾性检测,将耳念珠菌感染病例的时间前推至1996年,菌株是从当时一名患有吸入性肺炎的一岁女童的血液样本中鉴定出,这也是目前已知最早的耳念珠菌感染病例报告(Lee et al. 2011)。真菌耳乳突炎是一种很罕见的疾病,主要发生在免疫功能低下病人当中,多由曲霉引起,但随着耳念珠菌感染病例数目的增加,由耳念珠菌感染引起的耳乳突炎需要引起重视,2013年韩国报道了由耳念珠菌引起的耳乳突炎(Choi et al. 2017)。同年,印度德里两家医院的12个住院病人血液中分离出了耳念珠菌,随后,印度北部和南部卫生保健中心也相继报道耳念珠菌感染病例。据印度ICU全国调查显示,超过5%的念珠菌血症由耳念珠菌造成(Chowdhary et al. 2013;Chowdhary et al. 2014;Rudramurthy et al. 2017)。2017年巴基斯坦报道了首例耳念珠菌感染病例,随后报道的病例数不断上升(Lockhart et al. 2017)。在我国,2018年报道了首例耳念珠菌感染病例,系从北京大学人民医院一位76岁伴有基础病的病人支气管肺泡灌洗液中分离得到,该菌对抗真菌药物展现出敏感性特征(Wang et al. 2018)。随后,北京又报道了2例(Chen et al. 2018),沈阳通过回顾性研究报道了15例,样本来源有尿液、痰液、导管等(Tian et al. 2018),中国台湾地区也报道了1例(Tang et al. 2019),这些耳念珠菌对一种或几种抗真菌药具有抗性。在2012-2017年间,新加坡一家三级保健医院出现3位病人被耳念珠菌感染(Tan & Tan 2018)。2018年,马来西亚首次报道从一位患念珠菌血症和中性粒细胞减少症的63岁病人血液中检测到耳念珠菌和热带念珠菌(Mohd Tap et al. 2018)。此外,以色列(Ben-Ami et al. 2017)、科威特(Khan et al. 2018)、阿曼(Mohsin et al. 2017)、阿联酋(Alatoom et al. 2018)、沙特阿拉伯(Abdalhamid et al. 2018)、伊朗(Abastabar et al. 2019;Chow et al. 2019)等国也出现了耳念珠菌感染病例。
1.2 欧洲
在欧洲,已经有13个国家报道了耳念珠菌感染。2016年,西班牙报道了欧洲大陆首次出现的4例由耳念珠菌引起的念珠菌血症(Ruiz Gaitan et al. 2017)。同年,英国报道了英国皇家布朗普顿医院集中暴发的耳念珠菌感染,通过对2015年4月至2016年7月期间在院患者进行检测,共发现了50例患者被耳念珠菌感染(Schelenz et al. 2016)。此外,2015年2月至2017年8月期间,在牛津大学医院也发生了院内暴发性感染,共检出70例感染耳念珠菌的患者(Eyre et al. 2018)。正是这些集中性院内感染事件向全球卫生系统发出警告,引起各国的重视。从2018年1月到2019年5月,欧盟共有349例耳念珠菌感染病例(Plachouras et al. 2020),其中奥地利报道了一例耳念珠菌病例,系一位22岁患有外耳道感染的病人(Pekard-Amenitsch et al. 2018)。同年,在瑞士,从一位74岁患有急性呼吸窘迫综合症的女性患者的气道吸出物中分离得到了第一例耳念珠菌(Riat et al. 2018)。而在比利时,2018年报道的第一例耳念珠菌感染病例系来自科威特一名患有导管相关的念珠菌血症的女性患者(Dewaele et al. 2018)。此外,法国(Kohlenberg et al. 2018)、德国(Hamprecht et al. 2019)、挪威(Kohlenberg et al. 2018;Plachouras et al. 2020)、俄罗斯(Barantsevich et al. 2019)、希腊(Stathi et al. 2019)、意大利(Crea et al. 2019)、波兰和荷兰(Vogelzang et al. 2019)等国也相继报道耳念珠菌感染或携带病例。
1.3 非洲
非洲大陆属于热带气候,由于全球气候变暖、免疫功能低下患者增多以及发展随意性大,非洲大陆被称为传染病大陆。但令人惊讶的是,根据美国CDC的统计和文献检索,迄今为止整个非洲大陆报道耳念珠菌感染病例的国家并不多,目前仅有南非、肯尼亚、埃及和苏丹4个国家。其中,南非于2014年首次报道了4例耳念珠菌感染病例,患者发病时间为2012年10月——2013年10月,从这些念珠菌血症患者中分离到的菌株最开始均被错误地鉴定为希木龙念珠菌,它们对氟康唑均有耐药性(Magobo et al. 2014)。2016年,南非境内不同医院暴发了耳念珠菌感染。同年,在南非的私立和公立医院中,耳念珠菌分别被报道为第二和第四最常见的念珠菌血症诱因(Govender et al. 2018)。在肯尼亚,从2010年9月至2016年12月,38%的念珠菌血症是由耳念珠菌引起的,而最常见的白念珠菌引起的感染比例仅占25%,这些耳念珠菌起初也均被Vitek2错误地鉴定为希木龙念珠菌(Adam et al. 2019)。
1.4 南美洲
2016年,委内瑞拉首次报道了耳念珠菌引起的念珠菌血症感染。其第二大城市马拉开波的一家三级保健医院曾发生过聚集性感染,通过对2012年3月至2013年7月间ICU病房的18名患者血液中分离到的念珠菌进行重新鉴定发现,这些最初被鉴定为希木龙念珠菌的菌株其实是耳念珠菌,这些菌株对氟康唑均呈现出耐药性(Calvo et al. 2016)。自2013年起,哥伦比亚多个城市也出现耳念珠菌感染的散发病例(Parra-Giraldo et al. 2018)。
1.5 北美洲
在2013年5月至2016年8月间,美国首次发现7例耳念珠菌感染病例(Vallabhaneni et al. 2017)。截止2020年7月31日,CDC报道美国累计的耳念珠菌确诊病例已达到1 272例,此外还有30例疑似病例以及约2 493例耳念珠菌携带者。一项分子流行病学研究对2013年5月至2017年8月期间来自美国10个州的133例耳念珠菌感染病例进行调查,通过全基因组测序(WGS)分析后发现:这些临床分离株具有遗传多样性的特征,分别属于全球耳念珠菌的4大分支,其中约90%的菌株属于南亚分支(Clade I),7%的菌株属于南美分支(Clade IV),1%属于非洲分支(Clade III),1%属于东亚分支(Clade II),这说明耳念珠菌已被多次引进美国(Chow et al. 2018)。2017年5月,加拿大报道了首例多重耐药耳念珠菌(Schwartz & Hammond 2017)。而在2016年,巴拿马的一家医院从9位住院病人中分离出14株耳念珠菌(Arauz et al. 2018),这些分离株最初被Vitek 2自动系统鉴定为希木龙念珠菌,后来是通过分子方法确认为耳念珠菌。
1.6 大洋洲
2019年澳大利亚报道了首例耳念珠菌感染病例,这位65岁的男性患者曾于2012年在肯尼亚ICU接受治疗,2015年在澳大利亚被确诊为慢性耳念珠菌胸骨骨髓炎,该临床分离株属于南非分支(Heath et al. 2019)。
在上述40个报道耳念珠菌感染的国家中,很多是通过回顾性分析和再鉴定后发现的,因此,由于缺少现代实验室鉴定技术和检测范围的不足等客观因素的存在,那些未见报道的国家很可能也出现了耳念珠菌感染,特别是在一些检测技术手段较为落后的发展中国家。
2 耳念珠菌的形态学研究
2.1 菌丝发育
菌丝发育对真菌的致病性十分重要。真菌通过形成菌丝来侵袭宿主上皮细胞层,从而增强自身的毒性(Thompson et al. 2011)。细胞由酵母形态向菌丝形态转换是白念珠菌非常重要的生物学特征。白念珠菌菌丝的形成机理研究得较为深入,高温、碱性pH值、血清、CO2、GlcNAc等不同的环境条件均能诱导白念珠菌的菌丝发育(Huang 2012)。当耳念珠菌进入科学家的视线后,对其菌丝发育的研究也推到前沿。
最初的形态学研究发现,大多数耳念珠菌临床分离株均无法形成芽管、假菌丝、菌丝或厚垣孢子,即便是在白念珠菌菌丝诱导条件下(如高温、GlcNAc、血清),耳念珠菌细胞仍然维持着酵母形态(Wang et al. 2018)。但是,在某些特殊的培养环境下,一些临床菌株展现出菌丝生长的潜力。比如,在含10% NaCl的高盐压力培养条件下,耳念珠菌的细胞出现伸长的菌丝和假菌丝形态,但这些菌丝发育不完全(Wang et al. 2018);在含有吐温80的燕麦培养基上,耳念珠菌也能发育为假菌丝(Chew et al. 2019);在诱导生物膜形成时,少数耳念珠菌细胞也形成假菌丝(Sherry et al. 2017)。当用遗传毒性药物(比如羟基脲、甲磺酸甲酯、5-氟胞嘧啶)处理细胞后,耳念珠菌也能够形成假菌丝(Bravo Ruiz et al. 2020),不同来源的临床菌株对药物处理的应答程度不尽相同。因此,耳念珠菌菌丝形成的能力与遗传背景和培养环境均密切相关。此外研究表明,分子伴侣Hsp90参与调控耳念珠菌的形态发生,当细胞缺失HSP90或者用Hsp90抑制剂格尔德霉素处理细胞后,耳念珠菌能进行菌丝发育,说明HSP90是调控耳念珠菌菌丝发育的关键基因(Kim et al. 2019)。
目前虽然没有发现能够诱导耳念珠菌进行强菌丝发育的体外条件,但是经过哺乳动物宿主体内后,耳念珠菌却能进行由酵母态向菌丝态的可遗传的形态转换。将酵母形态的耳念珠菌经细胞尾静脉注入小鼠体内,从小鼠肾脏和肝脏分离出来的耳念珠菌在YPD培养基上培养24h后能形成非常细长的菌丝,这部分细胞能够长期保持菌丝发育的能力,耳念珠菌酵母形态和菌丝形态细胞分泌胞外蛋白酶的能力和在宿主不同器官的定植能力也明显不同。与白念珠菌的菌丝相比,耳念珠菌菌丝具有自身的特征:它通常具有多个液泡,并且并不十分规则,菌丝在低温25℃下比较稳定,而37℃则抑制菌丝的生长。转录组分析发现一些在白念珠菌菌丝形态细胞中表达上调的基因在耳念珠菌菌丝形态细胞中也高表达,如HGC1和ALS4;但也有一些基因(如EFG1)对白念珠菌菌丝发育至关重要,但在耳念珠菌菌丝细胞中的表达却下调(Yue et al. 2018)。比较基因组分析发现,耳念珠菌含有大多数参与白念珠菌菌丝调控基因的同源基因,但也有例外,如编码细胞溶解酶的ECE1和菌丝细胞壁蛋白的HWP1,它们在白念珠菌菌丝细胞中高表达,但在耳念珠菌基因组中缺失(Munoz et al. 2018)。这些都说明耳念珠菌与白念珠菌菌丝形成的机理有相同之处,也有自己独特的调控方式。
2.2 其他形态转换
除了菌丝发育,念珠菌属成员中还存在其他多种形态转换方式。比如在白念珠菌、热带念珠菌和都柏林念珠菌中均存在white、gray、opaque 3种细胞形态并且三者之间能进行可逆性地相互转换,从而形成一个三稳态转换系统,不同形态又可以相对稳定地进行遗传,转录因子Wor1在opaque形态形成过程中起关键作用(Yue et al. 2016;Zhang et al. 2016;Noble et al. 2017)。white形态是典型的酵母形态,在小鼠系统感染中毒性强于其他两种形态,opaque形态的细胞不仅与白念珠菌的毒性密切相关,它对皮肤的侵染能力较white细胞具有优势,还在有性生殖过程中起重要作用;gray形态的细胞在分泌胞外蛋白酶方面具有优势。这种形态可塑性使得细胞能定殖在不同的宿主生态位点。目前虽然在耳念珠菌中并未发现类似opaque和gray形态的细胞,但是它可能存在其他形式的形态转换。科玛嘉显色培养基常用于临床上念珠菌的鉴定,比如白念珠菌和热带念珠菌可以通过菌落颜色的变化来相对可靠地鉴定出,白念珠菌呈绿色,热带念珠菌呈蓝灰色。耳念珠菌和其他念珠菌如克柔假丝酵母Candida krusei、近平滑念珠菌Candida parapsilosis常呈现出粉色,不容易通过颜色区分开,临床常见的检测手段是用MALDI-TOF鉴定所有的粉色菌落,而其他非粉色的菌落常常被错误地排除在外。有报道显示耳念珠菌在科玛嘉显色培养基上会出现3种颜色的菌落:白色、粉色和深紫色,并且这3种菌落颜色的细胞能进行相互转换,类似于白念珠菌white-gray-opaque转换系统,但耳念珠菌不同颜色的菌落从细胞水平上看并无差别。耳念珠菌中存在3个与白念珠菌WOR1同源的基因(QG37_07829、CJI97_003997和CJJ07_002050),它们是否具有Wor1类似的功能还不清楚。
此外,部分耳念珠菌临床菌株在体外培养过程中会成产生成团现象,细胞出芽生殖后并不释放子细胞,导致大量细胞聚集在一起形成细胞团,这些聚集的细胞很难被打散,这种细胞被称为“聚集态细胞”,在大蜡螟毒性实验中,聚集态细胞的毒性比非聚集态细胞的毒性低(Borman et al. 2016)。在被耳念珠菌感染的小鼠肾脏中也发现了聚集态的细胞,细胞聚集化可能有利于耳念珠菌在宿主体内进行免疫逃逸和长期定殖于宿主体内(Ben-Ami et al. 2017)。
耳念珠菌的形态多样性及其转换的研究还处于起步阶段,这些不同形态在其致病性、毒性、有性生殖和宿主定殖能力中存在怎样的作用还有待进一步研究。
2.3 生物膜
生物膜是微生物细胞粘附在物体表面或者气液界面进行大量生长而产生的细胞群落,它是微生物常见的生存形式,在感染宿主的过程中至关重要。位于生物膜中的细胞不同于游离态的细胞,它们能更好地抵抗药物或者外界环境的影响,也能更好地逃避宿主免疫系统的攻击(Hall-Stoodley et al. 2004)。念珠菌生物膜是一个高度结构化的群体,在致病念珠菌中,白念珠菌形成生物膜的能力最强。典型的生物膜中含有酵母态、假菌丝态和菌丝态细胞,细胞包裹着大量的基质。生物膜不仅能在医疗器械、导管装置等处形成,还能在宿主表面(如口腔表面、上皮细胞内层等)形成,极大地增强了细胞对药物的耐受(Lohse et al. 2018),提升了白念珠菌的生存率和致死率(Rajendran et al. 2016)。相比白念珠菌而言,耳念珠菌形成生物膜的能力弱很多,但又明显比光滑念珠菌形成生物膜的能力强(Sherry et al. 2017)。
耳念珠菌的生物膜主要由芽殖酵母和极少数偶然出现的假菌丝所组成,胞外基质较少(Larkin et al. 2017);白念珠菌的生物膜则是由致密的菌丝和酵母细胞附着于细胞外基质上;光滑念珠菌形成的生物膜很稀薄,只由酵母细胞组成,并且没有细胞外基质。粘附素蛋白对白念珠菌粘附以及生物膜形成的起始至关重要,耳念珠菌基因组中粘附素家族基因数显著低于白念珠菌(Chatterjee et al. 2015;Munoz et al. 2018),但存在白念珠菌ALS1、ALS3和ALS5的同源基因(Kean et al. 2018;Singh et al. 2019),含有抗Als3抗体的血清能显著抑制耳念珠菌生物膜的形成,暗示Als3对耳念珠菌生物膜的形成必不可少(Singh et al. 2019)。转录组学分析发现,在耳念珠菌形成生物膜的整个过程中,粘附素相关的糖基磷脂酰肌醇(GPI)锚定的细胞壁基因CSA1、IFF4、PGA26和PGA52一直处于上调状态,而另外两个粘附素HYR3和ALS5仅在生物膜成熟阶段表达上调;当生物膜发生到中后期时,一系列编码外排泵的基因表达上调,包括ABC转运蛋白(CDR1、SNQ2和YHD3)和MFS转运蛋白(MDR1和RDC3),同时,当用外排泵抑制剂处理细胞后,耳念珠菌生物膜对氟康唑的敏感性增强2-8倍(Kean et al. 2018);而敲除CDR1后,所有原本对氟康唑耐药的临床菌株变得对氟康唑敏感,MIC值降低了128倍(Rybak et al. 2019)。因此,这些外排泵基因表达的上调在耳念珠菌耐药特征形成过程中起着重要作用。
虽然耳念珠菌生物膜的胞外基质较少,但其主要成分和白念珠菌等其他生物膜中类似,都是富含甘露聚糖-葡聚糖的多糖复合物,能将药物隔离在外从而阻止药物进入生物膜内部,对细胞起到很好的保护,导致整个菌体对药物产生耐受。耳念珠菌的胞外基质能将约70%的氟康唑隔离在胞外(Dominguez et al. 2019),耳念珠菌生物膜对卡泊芬净、米卡芬净和两性霉素B等抗真菌药物的敏感性更低(Sherry et al. 2017;Kean et al. 2018)。因此,耳念珠菌生物膜和其他念珠菌生物膜在功能上具有保守性,除了通过细胞外基质隔离药物外,还有多个机制参与其中:生物膜内细胞密度高;细胞生长受到营养限制,代谢缓慢;细胞中编码外排泵等抗性基因的表达上调;生物膜中存在“存留细胞”(Ramage et al. 2005)。
3 耳念珠菌的致病性与耐药研究
3.1 耳念珠菌的致病性
蛋白酶是目前最常见真菌毒性相关酶类。胞外水解酶的产生被认为是念珠菌属重要的毒性特征,与其致病性密切相关。此外,溶血素、脂酶和磷脂酶也起到重要作用。
分泌型天冬氨酸蛋白酶(SAPs)是白念珠菌产生的最有效的胞外酶之一。传统上认为,这些酶类通过降解宿主组织,从而为病原菌提供营养(Naglik et al. 2003)。在白念珠菌中,SAPs家族含有10个成员。其中Sap4、Sap5和Sap6对毒性起关键作用,抑制这些蛋白酶的产生将大大削弱白念珠菌的致病性(Lee et al. 2009)。致病念珠菌其他成员也有SAPs基因,包括都柏林念珠菌、近平滑念珠菌和热带念珠菌。基因组学分析表明热带念珠菌至少含有4个SAPs基因,近平滑念珠菌含有多达14个潜在的SAPs基因,都柏林念珠菌有8个天冬氨酸蛋白酶家族的成员。在耳念珠菌基因组(菌株编号Ci 6684,为印度来源)中,水解酶在所有酶类中占最大的比例(42%),发现有4个SAPs的同源基因,两个液泡型天冬氨酸蛋白酶(Chatterjee et al. 2015)。体外研究证实耳念珠菌能分泌蛋白酶,但分泌能力与菌株来源背景有关。比较耳念珠菌与白念珠菌的SAP活性发现,它们在25℃、37℃和40℃均具有较高活性。但耳念珠菌在42℃的SAP活性明显高于白念珠菌,暗示耳念珠菌在较高温度下仍能维持致病性(Wang et al. 2018)。
其他分子如溶血素也是病原菌定植宿主所必须的,许多常见的致病念珠菌均具有溶血素分泌活性,包括白念珠菌、都柏林念珠菌、光滑念珠菌和热带念珠菌。溶血素能帮助白念珠菌从宿主体内摄取铁,促进菌丝发育和侵入生长,进而提升致病率(Tsang et al. 2007)。从临床分离到的菌株比从环境来源的菌株具有更强的溶血素产生能力,说明溶血素也是一个重要的毒力因子。已有报告显示耳念珠菌也能产生溶血素(Kumar et al. 2015),但产溶血素能力是否与菌株来源有关并不清楚。
磷脂酶是病原菌分泌的另一毒性因子,它水解细胞膜主要成分磷脂,从而帮助病原菌进行侵入生长,同时在形成生物膜和免疫逃避的过程中也起重要作用(Samaranayake et al. 2006)。研究报道耳念珠菌也能分泌磷脂酶,37.5%的受检临床菌株均能产生磷脂酶,但产生磷脂酶的能力普遍弱于白念珠菌,且强弱与菌株来源有关(Kumar et al. 2015;Larkin et al. 2017)。但磷脂酶与耳念珠菌的毒性和致病性的相关性还需要进一步的实验证据。
3.2 耳念珠菌的耐药特征
耳念珠菌除了产生上述的毒性因子增强其致病性外,另一个导致感染的重要原因是其对药物的耐受,约90%的临床菌株对氟康唑具有很高的耐药性,因此很难被药物清除。这种耐药性的产生除了前述的形成生物膜以及相关外排泵基因上调之外,还包括一些参与物质合成途径的基因发生了突变。白念珠菌耐药性的产生原因之一是编码羊毛甾醇脱甲基酶的基因ERG11发生突变,或者ERG11表达上升,以及由于甾醇生物合成途径的改变而导致细胞质膜上的麦角甾醇被别的甾醇所替换(Cowen et al. 2014)。耳念珠菌也采用同样的策略,在对54株来源于4个分支的临床菌株进行全基因组分析发现,所有菌株的Erg11均发生了氨基酸突变,其中有3个突变热点,并且这3个突变热点与地理分支存在相关性:南非菌株为F126L,委内瑞拉菌株为Y132F,印度和巴基斯坦来源的菌株则为Y132F和K143R,这说明不同地理起源耳念珠菌的耐药性是进化获得而非固有的(Lockhart et al. 2017)。虽然和白念珠菌一样,ERG11的突变与氟康唑耐药性相关,但在氟康唑耐受菌株和敏感菌株中,ERG11的表达水平却没有显著性变化,这一点与白念珠菌不同。同样类似于白念珠菌棘白霉素耐受与FKS1基因突变相关,在所测试的耐受棘白霉素的耳念珠菌临床菌株中,Fks1均发生了S639F的突变(Chowdhary et al. 2018)。这些都说明耳念珠菌的致病策略与耐药策略与白念珠菌存在一定的保守性。
4 耳念珠菌的检测技术
4.1 基于培养与形态观察的方法
耳念珠菌在25-42℃条件下均能生长,在常用标准真菌培养基沙保氏琼脂培养基上,耳念珠菌呈现白色或米色、光滑、奶油状的菌落。临床上常用显色培养基对念珠菌进行培养,通过菌落颜色的差异来鉴定念珠菌成员,这种方法便捷快速。比如在常用的科玛嘉显色培养基上,白念珠菌呈现出绿色的菌落,热带念珠菌为蓝灰色菌落,光滑念珠菌呈现出紫色菌落,但在科玛嘉显色培养基,耳念珠菌的颜色并不典型,会形成白色、粉色和深紫色的菌落(Bentz et al. 2018),很难凭借颜色进行区分。但是,在添加有Pal’s(葵花籽提取)琼脂的科玛嘉显色培养基上培养,并结合耳念珠菌能在42℃的高温条件下也能很好地生长的特征,能快速高效地将耳念珠菌与其近亲希木龙念珠菌复合体进行区分,该方法价廉快速,其缺点是需要先用其他方法如VITEK2进行初步识别,先排除在科玛嘉显色培养基上形成粉色菌落的非白念珠菌成员(Kumar et al. 2017)。
除了耐受高温,耳念珠菌还具有耐高盐的特征,Welsh et al.(2017)利用此特征设计了两个能方便又快速的检测耳念珠菌的价廉培养基,他们在沙保氏培养基和酵母氮源培养基YNB中添加10% NaCl,并添加半乳糖醇和甘露醇作为碳源,在40℃条件下培养后,只有耳念珠菌能在此条件下生长,其他念珠菌包括其近亲希木龙念珠菌均不能在其生长。但在以葡萄糖为碳源的沙保氏培养基上,光滑念珠菌也能生长。
4.2 基质辅助激光解析离子飞行质谱(MALDI-TOF MS)
培养观察的方法虽然在改进后的培养基上能够较为准确地对耳念珠菌进行鉴定,但其用时相对较长,而且样本的处理量有限。近年来蛋白质组学的方法运用越来越广泛。其中MALDI-TOF MS已经进入临床微生物实验室用于病原菌鉴定。在临床微生物学实验室使用最广泛MALDI-TOF平台分别是德国的Bruker BioTyper系统和法国的Vitek MS系统。该方法是通过对细胞的蛋白质组进行检测,形成蛋白光谱,通过与数据库进行比对,找出与之相对应的物种。在数据库更新之前,耳念珠菌常常被MALDI-TOF MS错误地鉴定为其他念珠菌(Jeffery-Smith et al. 2018),随着数据库的更新,该方法的准确性得到极大地提升,甚至能对耳念珠菌进行基因分型(Prakash et al. 2016),MALDI-TOF MS未来还可能用于区分耐药菌与敏感菌株。该方法高效迅速,但需要购买特殊的仪器并不断更新数据库,并且菌株的培养条件和蛋白质的提取方法也会影响鉴定的准确性(Hata et al. 2020)。
4.3 基于PCR的分子鉴定和全基因组测序(WGS)
传统分子鉴定方法是通过PCR扩增ITS或D1/D2区域,将扩增产物测序后进行DNA序列比对,该方法是物种鉴定的金标准。通过序列分析,不仅可以鉴定出耳念珠菌,还能绘制进化树,将不同地域和不同来源的临床菌株进行深入分组。此外,通过优化PCR扩增引物,运用普通PCR或者荧光定量PCR进行扩增后,仅仅进行电泳或者溶解曲线分析,也能100%准确地鉴定耳念珠菌,鉴定所需的时间也缩短至2-2.5h(Kordalewska et al. 2017)。其他基于PCR反应的分型手段如扩增片段长度多样性(AFLP)和多位点序列分型(MLST)也可用于耳念珠菌的鉴定(Prakash et al. 2016)。
随着测序技术的进步,全基因组测序(WGS)越来越多地运用于对耳念珠菌进行分型和进化分析(Sharma et al. 2016;Lockhart et al. 2017)。由于其高分辨率,所以能比其他任何技术更好地进行进化和流行病学分析。缺点是价格昂贵,需要具备生物信息学处理能力。
5 耳念珠菌感染的防控
由于对耳念珠菌认识有限,人们起初并没有意识到需要对念珠菌成员采取接触防范措施。越来越多的证据显示耳念珠菌具有超强的体外生存能力,比如它能在地板、病床等干燥表面或者潮湿表面长时间生存(Piedrahita et al. 2017),耳念珠菌在干燥的塑料表面能至少存活14d,即便是28d后,也能检测到较高的酯酶活性(Welsh et al. 2017),表明耳念珠菌可在医院环境中定植,并通过卫生保健设施导致人传人(Piedrahita et al. 2017;Welsh et al. 2017;Kean et al. 2018;Sabino et al. 2020)。美国CDC已经发布了关于耳念珠菌感染控制措施的指南。耳念珠菌感染患者应当安置在单独的房间,并注意接触防范。看护耳念珠菌感染患者的医护人员需注重严格的手部卫生措施,用含酒精的洗手液或香皂认真清洗。此外,每天必须对患者的房间进行清洁和消毒,在别的患者入住前还要进行终端消毒。同时,应对所有设备进行严格规范的清洁消毒,并且正确处置废物和织布。对新感染患者的密切接触者需要进行耳念珠菌定殖筛查,筛查部位包括腋窝、腹股沟和鼻孔等处(Sexton et al. 2018;Wickes 2020)。
6 耳念珠菌研究的展望
作为一个新兴出现的病原真菌,耳念珠菌在全球迅速蔓延,对全球公共健康造成极大的威胁,但我们对它的认识还处于起步阶段。虽然从进化树分析可以推测耳念珠菌在不同地理位置是独立进化的,并且从各个不同来源的临床样本中均检测到耳念珠菌,但目前并不清楚它的生态起源是什么。此外,还需要加强耳念珠菌检测手段的开发,虽然MALDI-TOF MS、PCR和WGS等技术手段均能准确鉴定耳念珠菌,但这些方法目前并不能在世界上所有临床实验室开展,尤其是一些发展中国家,因此仍然有必要开发价廉、准确和高效的检测手段,以加大全世界耳念珠菌的监测力度。耳念珠菌在外部环境中能较长时间的生存,可持久定殖在医疗保健设施和人体宿主中,容易导致院内感染,因此,医疗机构需要制定并执行严格规范的卫生消毒程序。
耳念珠菌多重耐药的特征使得治疗方式有限,对药物的使用需要十分谨慎,目前可用的药物不多,棘白菌素类药物被认为是初始治疗的最佳药物。由于耳念珠菌的鉴定存在一定困难,经常被误检,这也给临床用药增加了难度,这种多重耐药特性的产生是否与临床用药存在关联还并不清楚。整体而言,耳念珠菌的耐药机理与研究较为深入的白念珠菌存在一定保守性,但它又有自己的特征,比如ERG11的突变热点在各个地理分支中存在特殊性;它形成生物膜和菌丝的能力没有白念珠菌强。因此对于耳念珠菌耐药机理还需要深入研究。此外,耳念珠菌是否存在有性生殖过程目前并不清楚,比较基因组分析发现,耳念珠菌含有大多数念珠菌交配相关的同源基因,隶属不同地理分支的成员在交配位点上具有偏好性,比如:Clade I(南亚分支)和Clade IV(南美分支)均为MTLa;Clade II(东亚分支)和Clade III(南非分支)均为MTLα(Munoz et al. 2018)。有性生殖对于物种毒力和环境适应能力的进化都非常重要,因此这也将是耳念珠菌研究的一大重点。
参考文献
Candida auris otomycosis in Iran and review of recent literature
DOI:10.1111/myc.12886
URL
PMID:30585653
[本文引用: 1]

Candida auris is a multidrug-resistant yeast emerging in immunocompromised and in otherwise healthy individuals. Due to difficulties in microbiological identification of C. auris because of the lack of available laboratory technology in developing countries, the number of patients affected is most likely underestimated. We report the first case of C. auris otitis which now adds Iran as the fifth country around the Persian Gulf, in addition to Kuwait, Oman, United Arab Emirates and Saudi Arabia. Candida auris is an unknown pathogen in routine laboratories in Iran because most Candida isolates are probably misdiagnosed. Otomycosis seems to be a different clinical presentation of C. auris mainly involving isolates from the East-Asian clade. We compared the mycological and clinical details of the Iranian patient with other cases of otitis reported since the last review of C. auris otomycosis in 2017.
First report of Candida auris infections from Saudi Arabia
DOI:10.1016/j.jiph.2018.05.010 URL PMID:29895475 [本文引用: 1]
In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris
Analysis of Candida auris fungemia at a single facility in Kenya
DOI:10.1016/j.ijid.2019.06.001
URL
PMID:31185293
[本文引用: 1]

OBJECTIVES: Candida auris emerged as a human pathogen in 2009 and has subsequently been identified around the world as a cause of invasive candidiasis. We did an analysis from a single institution in order to analyze risk factors and outcomes for C. auris candidemia. METHODS: Patients with candidemia were identified by the electronic medical record and reviewed for risk factors and outcome. Candida isolates were identified by Vitek2 as Candida haemulonii, but species determinations for 21 of the isolates using published molecular and proteomic methods identified all as C. auris. FINDINGS: From September 2010 to December 2016, C. auris accounted for 38% of 201 patients with candidemia, while C. albicans contributed 25%. C. auris patients had been hospitalized longer (mean 32 days vs. 13 days; p<0.001), were more likely to have central lines preceding candidemia than C. albicans patients (84% vs. 54%; p=<0.001) and had more commonly been treated with carbapenems (83% vs 61% for C. albicans [p=0.01]). The crude mortality was 29%, compared to 36% for C. albicans. CONCLUSIONS: These findings suggest an opportunistic pathogen that may be less virulent, but difficult to eradicate and that control efforts should focus on antimicrobial usage.
Persistent candidemia despite appropriate fungal therapy: first case of Candida auris from the United Arab Emirates
Isolation of Candida auris from 9 patients in Central America: importance of accurate diagnosis and susceptibility testing
DOI:10.1111/myc.12709
URL
PMID:28945325
[本文引用: 1]

Candida auris is an emerging multidrug-resistant (MDR) fungus associated with invasive infections and high mortality. This report describes 9 patients from whom C. auris was isolated at a hospital in Panama City, Panama, the first such cases in Central America, and highlights the challenges of accurate identification and methods for susceptibility testing.
Emergence of Candida auris in Russia
DOI:10.1016/j.jhin.2019.02.021
URL
PMID:30851375
[本文引用: 1]

This paper reports the emergence of Candida auris infections in an intensive care unit at a hospital in Moscow. Forty-nine cases were diagnosed in 2016-2017, and the risk factors and antifungal susceptibilities are described. The 30-day all-cause mortality for 19 bloodstream infections in patients who did not receive appropriate antifungal therapy was 42.1%. Phylogenetic analysis of the internal transcribed spacer and D1-D2 regions and K143R substitution in the ERG11 gene indicated that the studied C. auris strains were of South Asian origin. This first reported series of C. auris infections in Russia demonstrates the rapid dissemination of this species, and the need for international surveillance and control measures.
Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel
Phenotypic switching in newly emerged multidrug-resistant pathogen Candida auris
Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris
DOI:10.1016/j.diagmicrobio.2017.10.021
URL
PMID:29307565
[本文引用: 1]

CD101 is a new echinocandin with a prolonged half-life. CD101 was tested by broth microdilution against 100 isolates of the emerging yeast Candida auris. It showed activity against most isolates, including some that were resistant to other echinocandins.
Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species
Pseudohyphal growth of the emerging pathogen Candida auris is triggered by genotoxic stress through the S phase checkpoint
Hidden killers: human fungal infections
First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia
Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris
DOI:10.1186/s12864-015-1863-z
URL
PMID:26346253
[本文引用: 2]

BACKGROUND: Candida auris is a multidrug resistant, emerging agent of fungemia in humans. Its actual global distribution remains obscure as the current commercial methods of clinical diagnosis misidentify it as C. haemulonii. Here we report the first draft genome of C. auris to explore the genomic basis of virulence and unique differences that could be employed for differential diagnosis. RESULTS: More than 99.5 % of the C. auris genomic reads did not align to the current whole (or draft) genome sequences of Candida albicans, Candida lusitaniae, Candida glabrata and Saccharomyces cerevisiae; thereby indicating its divergence from the active Candida clade. The genome spans around 12.49 Mb with 8527 predicted genes. Functional annotation revealed that among the sequenced Candida species, it is closest to the hemiascomycete species Clavispora lusitaniae. Comparison with the well-studied species Candida albicans showed that it shares significant virulence attributes with other pathogenic Candida species such as oligopeptide transporters, mannosyl transfersases, secreted proteases and genes involved in biofilm formation. We also identified a plethora of transporters belonging to the ABC and major facilitator superfamily along with known MDR transcription factors which explained its high tolerance to antifungal drugs. CONCLUSIONS: Our study emphasizes an urgent need for accurate fungal screening methods such as PCR and electrophoretic karyotyping to ensure proper management of fungemia. Our work highlights the potential genetic mechanisms involved in virulence and pathogenicity of an important emerging human pathogen namely C. auris. Owing to its diversity at the genomic scale; we expect the genome sequence to be a useful resource to map species specific differences that will help develop accurate diagnostic markers and better drug targets.
Emergency of fungemia cases caused by fluconazole-resistant Candida auris in Beijing, China
Candida auris arriving on our shores: an Australian microbiology laboratory’s experience
Otomastoiditis caused by Candida auris: case report and literature review
DOI:10.1111/myc.12617
URL
PMID:28378904
[本文引用: 1]

Fungal otomastoiditis is a rare disease, but can be fatal for immunocompromised patients. Recently, there have been increasing cases of otologic infection caused by Candida auris. Candida auris can be easily misdiagnosed for other species and treatment is difficult due to multidrug resistance. Clinician should be aware of this rare pathogen, and it should be treated with appropriate antifungal agent with surgical debridement.
Potential fifth clade of Candida auris, Iran, 2018
DOI:10.3201/eid2509.190686
URL
PMID:31310230
[本文引用: 1]

Four major clades of Candida auris have been described, and all infections have clustered in these 4 clades. We identified an isolate representative of a potential fifth clade, separated from the other clades by >200,000 single-nucleotide polymorphisms, in a patient in Iran who had never traveled outside the country.
Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey.
DOI:10.1016/S1473-3099(18)30597-8
URL
PMID:30293877

BACKGROUND: Transmission of multidrug-resistant Candida auris infection has been reported in the USA. To better understand its emergence and transmission dynamics and to guide clinical and public health responses, we did a molecular epidemiological investigation of C auris cases in the USA. METHODS: In this molecular epidemiological survey, we used whole-genome sequencing to assess the genetic similarity between isolates collected from patients in ten US states (California, Connecticut, Florida, Illinois, Indiana, Maryland, Massachusetts, New Jersey, New York, and Oklahoma) and those identified in several other countries (Colombia, India, Japan, Pakistan, South Africa, South Korea, and Venezuela). We worked with state health departments, who provided us with isolates for sequencing. These isolates of C auris were collected during the normal course of clinical care (clinical cases) or as part of contact investigations or point prevalence surveys (screening cases). We integrated data from standardised case report forms and contact investigations, including travel history and epidemiological links (ie, patients that had shared a room or ward with a patient with C auris). Genetic diversity of C auris within a patient, a facility, and a state were evaluated by pairwise differences in single-nucleotide polymorphisms (SNPs). FINDINGS: From May 11, 2013, to Aug 31, 2017, isolates that corresponded to 133 cases (73 clinical cases and 60 screening cases) were collected. Of 73 clinical cases, 66 (90%) cases involved isolates related to south Asian isolates, five (7%) cases were related to South American isolates, one (1%) case to African isolates, and one (1%) case to east Asian isolates. Most (60 [82%]) clinical cases were identified in New York and New Jersey; these isolates, although related to south Asian isolates, were genetically distinct. Genomic data corroborated five (7%) clinical cases in which patients probably acquired C auris through health-care exposures abroad. Among clinical and screening cases, the genetic diversity of C auris isolates within a person was similar to that within a facility during an outbreak (median SNP difference three SNPs, range 0-12). INTERPRETATION: Isolates of C auris in the USA were genetically related to those from four global regions, suggesting that C auris was introduced into the USA several times. The five travel-related cases are examples of how introductions can occur. Genetic diversity among isolates from the same patients, health-care facilities, and states indicates that there is local and ongoing transmission. FUNDING: US Centers for Disease Control and Prevention.
Multidrug-resistant endemic clonal strain of Candida auris in India
DOI:10.1007/s10096-013-2027-1
URL
PMID:24357342
[本文引用: 2]

Candida auris is a recently described rare agent of fungemia. It is notable for its antifungal resistance. A total of 15 C. auris isolates, originating from seven cases of fungemia, three cases of diabetic gangrenous foot, and one case of bronchopneumonia from a tertiary care hospital in south India, were investigated. All of the 15 isolates were identified by sequencing and 14 of these along with 12 C. auris isolates previously reported from two hospitals in Delhi, north India, two each from Japan and Korea were genotyped by amplified fragment length polymorphism (AFLP). In vitro antifungal susceptibility testing (AFST) was done by the Clinical and Laboratory Standards Institute (CLSI) broth microdilution method. Candida auris isolates were misidentified as Candida haemulonii by VITEK. All were resistant to fluconazole [geometric mean minimum inhibitory concentration (MIC) 64 mug/ml] and 11 isolates were resistant to voriconazole (MIC >/=1 mug/ml). Forty-seven percent of the C. auris isolates were resistant to flucytosine (MIC >/=64 mug/ml) and 40% had high MIC (>/=1 mug/ml) of caspofungin. Breakthrough fungemia developed in 28.6% of patients and therapeutic failure in 4 (66.7%) patients. Interestingly, the 26 Indian C. auris isolates from north and south India were clonal and phenotypically and genotypically distinct from Korean and Japanese isolates. The present study demonstrates that C. auris is a potential emerging pathogen that can cause a wide spectrum of human mycotic infections. The prevalence of a C. auris endemic clonal strain resistant to azoles and other antifungals in Indian hospitals with high rates of therapeutic failure in cases of fungemia is worrisome.
A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance
DOI:10.1093/jac/dkx480
URL
PMID:29325167
[本文引用: 2]

Background: Candida auris has emerged globally as an MDR nosocomial pathogen in ICU patients. Objectives: We studied the antifungal susceptibility of C. auris isolates (n = 350) from 10 hospitals in India collected over a period of 8 years. To investigate azole resistance, ERG11 gene sequencing and expression profiling was conducted. In addition, echinocandin resistance linked to mutations in the C. auris FKS1 gene was analysed. Methods: CLSI antifungal susceptibility testing of six azoles, amphotericin B, three echinocandins, terbinafine, 5-flucytosine and nystatin was conducted. Screening for amino acid substitutions in ERG11 and FKS1 was performed. Results: Overall, 90% of C. auris were fluconazole resistant (MICs 32 to >/=64 mg/L) and 2% and 8% were resistant to echinocandins (>/=8 mg/L) and amphotericin B (>/=2 mg/L), respectively. ERG11 sequences of C. auris exhibited amino acid substitutions Y132 and K143 in 77% (n = 34/44) of strains that were fluconazole resistant whereas WT genotypes, i.e. without substitutions at these positions, were observed in isolates with low fluconazole MICs (1-2 mg/L) suggesting that these substitutions confer a phenotype of resistance to fluconazole similar to that described for Candida albicans. No significant expression of ERG11 was observed, although expression was inducible in vitro with fluconazole exposure. Echinocandin resistance was linked to a novel mutation S639F in FKS1 hot spot region I. Conclusions: Overall, 25% and 13% of isolates were MDR and multi-azole resistant, respectively. The most common resistance combination was azoles and 5-flucytosine in 14% followed by azoles and amphotericin B in 7% and azoles and echinocandins in 2% of isolates.
New clonal strain of Candida auris, Delhi, India
Candida auris: a rapidly emerging cause of hospital-acquired multidrug- resistant fungal infections globally
Mechanisms of antifungal drug resistance
DOI:10.1101/cshperspect.a019752
URL
PMID:25384768
[本文引用: 1]

Antifungal therapy is a central component of patient management for acute and chronic mycoses. Yet, treatment choices are restricted because of the sparse number of antifungal drug classes. Clinical management of fungal diseases is further compromised by the emergence of antifungal drug resistance, which eliminates available drug classes as treatment options. Once considered a rare occurrence, antifungal drug resistance is on the rise in many high-risk medical centers. Most concerning is the evolution of multidrug- resistant organisms refractory to several different classes of antifungal agents, especially among common Candida species. The mechanisms responsible are mostly shared by both resistant strains displaying inherently reduced susceptibility and those acquiring resistance during therapy. The molecular mechanisms include altered drug affinity and target abundance, reduced intracellular drug levels caused by efflux pumps, and formation of biofilms. New insights into genetic factors regulating these mechanisms, as well as cellular factors important for stress adaptation, provide a foundation to better understand the emergence of antifungal drug resistance.
Isolation of Candida auris from invasive and non-invasive samples of a patient suffering from vascular disease, Italy, July 2019
First case of Candida auris infection in Belgium in a surgical patient from Kuwait
Conserved role for biofilm matrix polysaccharides in Candida auris drug resistance
Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance
A Candida auris outbreak and its control in an intensive care setting
DOI:10.1056/NEJMoa1714373
URL
PMID:30281988
[本文引用: 1]

BACKGROUND: Candida auris is an emerging and multidrug-resistant pathogen. Here we report the epidemiology of a hospital outbreak of C. auris colonization and infection. METHODS: After identification of a cluster of C. auris infections in the neurosciences intensive care unit (ICU) of the Oxford University Hospitals, United Kingdom, we instituted an intensive patient and environmental screening program and package of interventions. Multivariable logistic regression was used to identify predictors of C. auris colonization and infection. Isolates from patients and from the environment were analyzed by whole-genome sequencing. RESULTS: A total of 70 patients were identified as being colonized or infected with C. auris between February 2, 2015, and August 31, 2017; of these patients, 66 (94%) had been admitted to the neurosciences ICU before diagnosis. Invasive C. auris infections developed in 7 patients. When length of stay in the neurosciences ICU and patient vital signs and laboratory results were controlled for, the predictors of C. auris colonization or infection included the use of reusable skin-surface axillary temperature probes (multivariable odds ratio, 6.80; 95% confidence interval [CI], 2.96 to 15.63; P<0.001) and systemic fluconazole exposure (multivariable odds ratio, 10.34; 95% CI, 1.64 to 65.18; P=0.01). C. auris was rarely detected in the general environment. However, it was detected in isolates from reusable equipment, including multiple axillary skin-surface temperature probes. Despite a bundle of infection-control interventions, the incidence of new cases was reduced only after removal of the temperature probes. All outbreak sequences formed a single genetic cluster within the C. auris South African clade. The sequenced isolates from reusable equipment were genetically related to isolates from the patients. CONCLUSIONS: The transmission of C. auris in this hospital outbreak was found to be linked to reusable axillary temperature probes, indicating that this emerging pathogen can persist in the environment and be transmitted in health care settings. (Funded by the National Institute for Health Research Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance at Oxford University and others.).
Candida auris in South Africa, 2012-2016
DOI:10.3201/eid2411.180368
URL
PMID:30334713
[本文引用: 1]

To determine the epidemiology of Candida auris in South Africa, we reviewed data from public- and private-sector diagnostic laboratories that reported confirmed and probable cases of invasive disease and colonization for October 2012-November 2016. We defined a case as a first isolation of C. auris from any specimen from a person of any age admitted to any healthcare facility in South Africa. We defined probable cases as cases where the diagnostic laboratory had used a nonconfirmatory biochemical identification method and C. haemulonii was cultured. We analyzed 1,692 cases; 93% were from private-sector healthcare facilities, and 92% of cases from known locations were from Gauteng Province. Of cases with available data, 29% were invasive infections. The number of cases increased from 18 (October 2012-November 2013) to 861 (October 2015-November 2016). Our results show a large increase in C. auris cases during the study period, centered on private hospitals in Gauteng Province.
Bacterial biofilms: from the natural environment to infectious diseases.
DOI:10.1038/nrmicro821
URL
PMID:15040259
[本文引用: 1]

Biofilms--matrix-enclosed microbial accretions that adhere to biological or non-biological surfaces--represent a significant and incompletely understood mode of growth for bacteria. Biofilm formation appears early in the fossil record (approximately 3.25 billion years ago) and is common throughout a diverse range of organisms in both the Archaea and Bacteria lineages, including the 'living fossils' in the most deeply dividing branches of the phylogenetic tree. It is evident that biofilm formation is an ancient and integral component of the prokaryotic life cycle, and is a key factor for survival in diverse environments. Recent advances show that biofilms are structurally complex, dynamic systems with attributes of both primordial multicellular organisms and multifaceted ecosystems. Biofilm formation represents a protected mode of growth that allows cells to survive in hostile environments and also disperse to colonize new niches. The implications of these survival and propagative mechanisms in the context of both the natural environment and infectious diseases are discussed in this review.
Candida auris in Germany and previous exposure to foreign healthcare
DOI:10.3201/eid2509.190262
URL
PMID:31223105
[本文引用: 1]

The emerging yeast Candida auris has disseminated worldwide. We report on 7 cases identified in Germany during 2015-2017. In 6 of these cases, C. auris was isolated from patients previously hospitalized abroad. Whole-genome sequencing and epidemiologic analyses revealed that all patients in Germany were infected with different strains.
Candida auris: an emerging yeast pathogen posing distinct challenges for laboratory diagnostics, treatment, and infection prevention
DOI:10.5858/arpa.2018-0508-RA
URL
PMID:31169997
[本文引用: 1]

CONTEXT.-: Candida auris is an emerging yeast species that was first described in 2009. This ascomycetous yeast is notable for resistance to azole antifungal agents, for environmental persistence, and for its ability to contaminate health care environments, resulting in patient colonization and nosocomial infections. OBJECTIVE.-: To review the state of current knowledge addressing challenges in the accurate identification of C auris in the diagnostic microbiology laboratory, including application of phenotypic, proteomic, and genomic methodologies; characteristics that may predispose the human host to acquiring C auris; transmission; clinical presentations; treatment modalities; environmental decontamination; and infection prevention in health care settings. DATA SOURCES.-: The PubMed search engine was used to access peer-reviewed literature published from 2009 to 2019. CONCLUSIONS.-: The rapid emergence of C auris has presented unique challenges for the areas of laboratory diagnostics and infection prevention and in options for antifungal treatment, which are limited. The current lack of established antifungal susceptibility test breakpoints complicates therapeutic decision making. Enhanced awareness of this pathogen is essential to monitor outbreaks and to reduce the risk of spread within health care environments.
Candida auris sternal osteomyelitis in a man from Kenya visiting Australia, 2015
DOI:10.3201/eid2501.181321
URL
PMID:30561310
[本文引用: 1]

In Australia in 2015, Candida auris sternal osteomyelitis was diagnosed in a 65-year-old man with a history of intensive care treatment in Kenya in 2012 and without a history of cardiac surgery. The isolate was South Africa clade III. Clinicians should note that C. auris can cause low-grade disease years after colonization.
Regulation of phenotypic transitions in the fungal pathogen Candida albicans
DOI:10.4161/viru.20010
URL
PMID:22546903
[本文引用: 1]

The human commensal fungus Candida albicans can cause not only superficial infections, but also life-threatening disease in immunocompromised individuals. C. albicans can grow in several morphological forms. The ability to switch between different phenotypic forms has been thought to contribute to its virulence. The yeast-filamentous growth transition and white-opaque switching represent two typical morphological switching systems, which have been intensively studied in C. albicans. The interplay between environmental factors and genes determines the morphology of C. albicans. This review focuses on the regulation of phenotypic changes in this pathogenic organism by external environmental cues and internal genes.
Candida auris: a review of the literature
DOI:10.1128/CMR.00029-17
URL
PMID:29142078
[本文引用: 1]

The emerging pathogen Candida auris has been associated with nosocomial outbreaks on five continents. Genetic analysis indicates the simultaneous emergence of separate clades of this organism in different geographical locations. Invasive infection and colonization have been detected predominantly in patients in high-dependency settings and have garnered attention due to variable antifungal resistance profiles and transmission within units instituting a range of infection prevention and control measures. Issues with the identification of C. auris using both phenotypic and molecular techniques have raised concerns about detecting the true scale of the problem. This review considers the literature available on C. auris and highlights the key unknowns, which will provide direction for further work in this field.
Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption Ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method
DOI:10.1128/JCM.00367-15
URL
PMID:25809970
[本文引用: 1]

Candida auris is a multidrug-resistant yeast that causes a wide spectrum of infections, especially in intensive care settings. We investigated C. auris prevalence among 102 clinical isolates previously identified as Candida haemulonii or Candida famata by the Vitek 2 system. Internal transcribed spacer region (ITS) sequencing confirmed 88.2% of the isolates as C. auris, and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) easily separated all related species, viz., C. auris (n = 90), C. haemulonii (n = 6), C. haemulonii var. vulnera (n = 1), and Candida duobushaemulonii (n = 5). The in vitro antifungal susceptibility was determined using CLSI broth microdilution (CLSI-BMD), the Vitek 2 antifungal susceptibility test, and the Etest method. C. auris isolates revealed uniformly elevated fluconazole MICs (MIC50, 64 mug/ml), and an alarming percentage of isolates (37%) exhibited elevated caspofungin MICs by CLSI-BMD. Notably, 34% of C. auris isolates had coexisting elevated MICs (>/=2 mug/ml) for both fluconazole and voriconazole, and 10% of the isolates had elevated coexisting MICs (>/=2 mug/ml) to two additional azoles, i.e., posaconazole and isavuconazole. In contrast to reduced amphotericin B MICs by CLSI-BMD (MIC50, 1 mug/ml) for C. auris, elevated MICs were noted by Vitek 2 (MIC50, 8 mug/ml), which were statistically significant. Candida auris remains an unnoticed pathogen in routine microbiology laboratories, as 90% of the isolates characterized by commercial identification systems are misidentified as C. haemulonii. MALDI-TOF MS proved to be a more robust diagnostic technique for rapid identification of C. auris. Considering that misleading elevated MICs of amphotericin B by the Vitek AST-YS07 card may lead to the selection of inappropriate therapy, a cautionary approach is recommended for laboratories relying on commercial systems for identification and antifungal susceptibility testing of rare yeasts.
Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm- mediated resistance
DOI:10.1128/mSphere.00334-18
URL
PMID:29997121
[本文引用: 3]

Candida auris has emerged as a significant global nosocomial pathogen. This is primarily due to its antifungal resistance profile but also its capacity to form adherent biofilm communities on a range of clinically important substrates. While we have a comprehensive understanding of how other Candida species resist and respond to antifungal challenge within the sessile phenotype, our current understanding of C. auris biofilm-mediated resistance is lacking. In this study, we are the first to perform transcriptomic analysis of temporally developing C. auris biofilms, which were shown to exhibit phase- and antifungal class-dependent resistance profiles. A de novo transcriptome assembly was performed, where sequenced sample reads were assembled into an ~11.5-Mb transcriptome consisting of 5,848 genes. Differential expression (DE) analysis demonstrated that 791 and 464 genes were upregulated in biofilm formation and planktonic cells, respectively, with a minimum 2-fold change. Adhesin-related glycosylphosphatidylinositol (GPI)-anchored cell wall genes were upregulated at all time points of biofilm formation. As the biofilm developed into intermediate and mature stages, a number of genes encoding efflux pumps were upregulated, including ATP-binding cassette (ABC) and major facilitator superfamily (MFS) transporters. When we assessed efflux pump activity biochemically, biofilm efflux was greater than that of planktonic cells at 12 and 24 h. When these were inhibited, fluconazole sensitivity was enhanced 4- to 16-fold. This study demonstrates the importance of efflux-mediated resistance within complex C. auris communities and may explain the resistance of C. auris to a range of antimicrobial agents within the hospital environment.IMPORTANCE Fungal infections represent an important cause of human morbidity and mortality, particularly if the fungi adhere to and grow on both biological and inanimate surfaces as communities of cells (biofilms). Recently, a previously unrecognized yeast, Candida auris, has emerged globally that has led to widespread concern due to the difficulty in treating it with existing antifungal agents. Alarmingly, it is also able to grow as a biofilm that is highly resistant to antifungal agents, yet we are unclear about how it does this. Here, we used a molecular approach to investigate the genes that are important in causing the cells to be resistant within the biofilm. The work provides significant insights into the importance of efflux pumps, which actively pump out toxic antifungal drugs and therefore enhance fungal survival within a variety of harsh environments.
Surface disinfection challenges for Candida auris: an in-vitro study
DOI:10.1016/j.jhin.2017.11.015
URL
PMID:29203448
[本文引用: 1]

The emerging pathogenic multidrug-resistant yeast Candida auris is an important source of healthcare-associated infections and of growing global clinical concern. The ability of this organism to survive on surfaces and withstand environmental stressors creates a challenge for eradicating it from hospitals. A panel of C. auris clinical isolates was evaluated on different surface environments against the standard disinfectant sodium hypochlorite and high-level disinfectant peracetic acid. C. auris was shown to selectively tolerate clinically relevant concentrations of sodium hypochlorite and peracetic acid in a surface-dependent manner, which may explain its ability to successfully persist within the hospital environment.
Invasive Candida auris infections in Kuwait hospitals: epidemiology, antifungal treatment and outcome
DOI:10.1007/s15010-018-1164-y
URL
PMID:29949089
[本文引用: 1]

PURPOSE: Candida auris is a recently recognized yeast pathogen, which has attracted worldwide attention due to its multidrug-resistant nature and associated high mortality rates. Its persistence in hospital environment and propensity of nosocomial transmission underscores the need of continuous monitoring to prevent outbreaks. Since the first case of C. auris candidemia in May, 2014, we have identified 17 additional invasive cases, which are described here. METHODS: Identity of 17 isolates originating from proven or possible cases of invasive C. auris infection and identified as Candida haemulonii by Vitek 2 yeast identification system was confirmed by PCR-sequencing of rDNA. Information about risk factors, treatment and outcomes were retrospectively retrieved from case files. Antifungal susceptibility testing was performed by Etest. RESULTS: Thirteen cases of candidemia and 4 cases of other invasive infections were detected in 6 hospitals across Kuwait. Major risk factors included adult patients with cancer, diabetes, gastrointestinal/liver diseases and extended (> 25 days) hospital stay. All isolates were resistant to fluconazole. Additionally, 5 and 4 isolates were also resistant to voriconazole and amphotericin B, respectively. Despite antifungal treatment, 9 of 15 patients died. Most patients (n = 12) were hospitalized in 2 hospitals that are in close proximity, whereas 5 other patients were from 3 hospitals that are situated > 10 km apart. CONCLUSIONS: Occurrence of successive cases of invasive C. auris infections with resulting mortality in nine patients suggests persistence of this multidrug-resistant yeast in major hospitals in Kuwait. Early detection by continuous surveillance and enforcement of infection control measures are recommended.
Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features
Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance
DOI:10.1128/mBio.02529-18
URL
PMID:30696744
[本文引用: 1]

Candida auris is an emerging fungal pathogen and a serious global health threat as the majority of clinical isolates display elevated resistance to currently available antifungal drugs. Despite the increased prevalence of C. auris infections, the mechanisms governing drug resistance remain largely elusive. In diverse fungi, the evolution of drug resistance is enabled by the essential molecular chaperone Hsp90, which stabilizes key regulators of cellular responses to drug-induced stress. Hsp90 also orchestrates temperature-dependent morphogenesis in Candida albicans, a key virulence trait. However, the role of Hsp90 in the pathobiology of C. auris remains unknown. In order to study regulatory functions of Hsp90 in C. auris, we placed HSP90 under the control of a doxycycline-repressible promoter to enable transcriptional repression. We found that Hsp90 is essential for growth in C. auris and that it enables tolerance of clinical isolates with respect to the azoles, which inhibit biosynthesis of the membrane sterol ergosterol. High-level azole resistance was independent of Hsp90 but dependent on the ABC transporter CDR1, deletion of which resulted in abrogated resistance. Strikingly, we discovered that C. auris undergoes a morphogenetic transition from yeast to filamentous growth in response to HSP90 depletion or cell cycle arrest but not in response to other cues that induce C. albicans filamentation. Finally, we observed that this developmental transition is associated with global transcriptional changes, including the induction of cell wall-related genes. Overall, this report provides a novel insight into mechanisms of drug tolerance and resistance in C. auris and describes a developmental transition in response to perturbation of a core regulator of protein homeostasis.IMPORTANCE Fungal pathogens pose a serious threat to public health. Candida auris is an emerging fungal pathogen that is often resistant to commonly used antifungal drugs. However, the mechanisms governing drug resistance and virulence in this organism remain largely unexplored. In this study, we adapted a conditional expression system to modulate the transcription of an essential gene, HSP90, which regulates antifungal resistance and virulence in diverse fungal pathogens. We showed that Hsp90 is essential for growth in C. auris and is important for tolerance of the clinically important azole antifungals, which block ergosterol biosynthesis. Further, we established that the Cdr1 efflux transporter regulates azole resistance. Finally, we discovered that C. auris transitions from yeast to filamentous growth in response to Hsp90 inhibition, accompanied by global transcriptional remodeling. Overall, this work provides a novel insight into mechanisms regulating azole resistance in C. auris and uncovers a distinct developmental program regulated by Hsp90.
Candida auris: epidemiological situation, laboratory capacity and preparedness in European Union and European Economic Area countries, 2013 to 2017.
Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris
Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal’s medium
DOI:10.1016/j.riam.2016.11.004 URL [本文引用: 1]
Itraconazole- resistant Candida auris with phospholipase, proteinase and hemolysin activity from a case of vulvovaginitis
DOI:10.3855/jidc.4582
URL
PMID:25881537
[本文引用: 2]

Since the emergence of pathogenic non-albicans Candida species, a number of new isolates have been added to the list. One such unusual species is Candida auris (C. auris), recently isolated and studied in few reports. In this study, a case of vulvovaginitis caused by Candida auris incidentally identified by molecular methods using internal transcribed spacer polymerase chain reaction (ITS PCR) is described. Antifungal susceptibility testing revealed the isolate to be resistant to itraconazole (MIC >/= 2 microg/ml) and expressed important virulence factors including phospholipase, proteinase and hemolysin activity. The patient was successfully treated with oral fluconazole and did not have any invasive fungemia. Very few cases of this emerging pathogen have been reported. However, its isolation from clinical specimens reveals the significance of non-albicans candida species over C. albicans and the diversity of Candida spp causing infections.
The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation
DOI:10.1128/AAC.02396-16
URL
PMID:28223375
[本文引用: 2]

Candidaauris, a new multidrug-resistant Candida spp. which is associated with invasive infection and high rates of mortality, has recently emerged. Here, we determined the virulence factors (germination, adherence, biofilm formation, phospholipase and proteinase production) of 16 C. auris isolates and their susceptibilities to 11 drugs belonging to different antifungal classes, including a novel orally bioavailable 1,3-beta-d-glucan synthesis inhibitor (SCY-078). We also examined the effect of SCY-078 on the growth, ultrastructure, and biofilm-forming abilities of C. auris Our data showed that while the tested strains did not germinate, they did produce phospholipase and proteinase in a strain-dependent manner and had a significantly reduced ability to adhere and form biofilms compared to that of Candida albicans (P = 0.01). C. auris isolates demonstrated reduced susceptibility to fluconazole and amphotericin B, while, in general, they were susceptible to the remaining drugs tested. SCY-078 had an MIC90 of 1 mg/liter against C. auris and caused complete inhibition of the growth of C. auris and C. albicans Scanning electron microscopy analysis showed that SCY-078 interrupted C. auris cell division, with the organism forming abnormal fused fungal cells. Additionally, SCY-078 possessed potent antibiofilm activity, wherein treated biofilms demonstrated significantly reduced metabolic activity and a significantly reduced thickness compared to the untreated control (P < 0.05 for both comparisons). Our study shows that C. auris expresses several virulence determinants (albeit to a lesser extent than C. albicans) and is resistant to fluconazole and amphotericin B. SCY-078, the new orally bioavailable antifungal, had potent antifungal/antibiofilm activity against C. auris, indicating that further evaluation of this antifungal is warranted.
Candida albicans VPS4 is required for secretion of aspartyl proteases and in vivo virulence
First three reported cases of nosocomial fungemia caused by Candida auris
DOI:10.1128/JCM.00319-11
URL
PMID:21715586
[本文引用: 1]

Candida auris is a newly described species whose clinical significance is not clear. Here, we describe the first three cases of nosocomial fungemia caused by C. auris, which confirms that it is a causative agent of bloodstream infections. All three patients presented persistent fungemia for 10 to 31 days. The isolates obtained from the three patients were misidentified as Candida haemulonii and Rhodotorula glutinis by the Vitek 2 and the API 20C systems, respectively. C. auris was confirmed by sequence analysis of the internal transcribed spacer region and D1/D2 regions of the 26S ribosomal DNA of the rRNA gene. The MIC ranges of amphotericin B (AMB), fluconazole (FLU), itraconazole, and voriconazole were 0.5 to 1, 2 to 128, 0.125 to 2, and 0.06 to 1 mug/ml, respectively. All isolates were susceptible to caspofungin (MIC = 0.06 mug/ml) and micafungin (MIC = 0.03 mug/ml). One patient developed breakthrough fungemia while receiving FLU therapy, and two patients who received FLU therapy followed by AMB showed therapeutic failure and fatal outcomes. Our cases show that C. auris fungemia can be persistent, despite FLU or AMB therapy, which emphasizes the importance of accurately identifying this species.
Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses
Development and regulation of single- and multi-species Candida albicans biofilms.
DOI:10.1038/nrmicro.2017.107
URL
PMID:29062072
[本文引用: 1]

Candida albicans is among the most prevalent fungal species of the human microbiota and asymptomatically colonizes healthy individuals. However, it is also an opportunistic pathogen that can cause severe, and often fatal, bloodstream infections. The medical impact of C. albicans typically depends on its ability to form biofilms, which are closely packed communities of cells that attach to surfaces, such as tissues and implanted medical devices. In this Review, we provide an overview of the processes involved in the formation of C. albicans biofilms and discuss the core transcriptional network that regulates biofilm development. We also consider some of the advantages that biofilms provide to C. albicans in comparison with planktonic growth and explore polymicrobial biofilms that are formed by C. albicans and certain bacterial species.
Candida auris-associated candidemia, South Africa
DOI:10.3201/eid2007.131765 URL PMID:24963796 [本文引用: 1]
A fatal case of Candida auris and Candida tropicalis candidemia in neutropenic patient
DOI:10.1007/s11046-018-0244-y
URL
PMID:29383574
[本文引用: 1]

We report a fatal case of Candida auris that was involved in mixed candidemia with Candida tropicalis, isolated from the blood of a neutropenic patient. Identification of both isolates was confirmed by amplification and sequencing of internal transcribed spacer and D1/D2 domain of large subunit in rRNA gene. Antifungal susceptibility test by E-test method revealed that C. auris was resistant to amphotericin B, anidulafungin, caspofungin, fluconazole, itraconazole and voriconazole. On the other hand, C. tropicalis was sensitive to all antifungal tested. The use of chromogenic agar as isolation media is vital in detecting mixed candidemia.
The first cases of Candida auris candidaemia in Oman
DOI:10.1111/myc.12647
URL
PMID:28685887
[本文引用: 1]

Candida auris has been recognised as a problematic healthcare-associated emerging yeast which is often misidentified as Candida haemulonii by commercial systems. Correct early identification of C. auris is important for appropriate antifungal treatment and implementing effective infection control measures. Here we report emergence of the first C. auris cases in Oman, initially misidentified as C. haemulonii.
Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species
DOI:10.1038/s41467-018-07779-6
URL
PMID:30559369
[本文引用: 3]

Candida auris is an emergent multidrug-resistant fungal pathogen causing increasing reports of outbreaks. While distantly related to C. albicans and C. glabrata, C. auris is closely related to rarely observed and often multidrug-resistant species from the C. haemulonii clade. Here, we analyze near complete genome assemblies for the four C. auris clades and three related species, and map intra- and inter-species rearrangements across the seven chromosomes. Using RNA-Seq-guided gene predictions, we find that most mating and meiosis genes are conserved and that clades contain either the MTLa or MTLalpha mating loci. Comparing the genomes of these emerging species to those of other Candida species identifies genes linked to drug resistance and virulence, including expanded families of transporters and lipases, as well as mutations and copy number variants in ERG11. Gene expression analysis identifies transporters and metabolic regulators specific to C. auris and those conserved with related species which may contribute to differences in drug response in this emerging fungal clade.
Candida albicans secreted aspartyl proteinases in virulence and pathogenesis
Candida albicans cell-type switching and functional plasticity in the mammalian host.
DOI:10.1038/nrmicro.2016.157
URL
PMID:27867199
[本文引用: 1]

Candida albicans is a ubiquitous commensal of the mammalian microbiome and the most prevalent fungal pathogen of humans. A cell-type transition between yeast and hyphal morphologies in C. albicans was thought to underlie much of the variation in virulence observed in different host tissues. However, novel yeast-like cell morphotypes, including opaque(a/alpha), grey and gastrointestinally induced transition (GUT) cell types, were recently reported that exhibit marked differences in vitro and in animal models of commensalism and disease. In this Review, we explore the characteristics of the classic cell types - yeast, hyphae, pseudohyphae and chlamydospores - as well as the newly identified yeast-like morphotypes. We highlight emerging knowledge about the associations of these different morphotypes with different host niches and virulence potential, as well as the environmental cues and signalling pathways that are involved in the morphological transitions.
First report of sporadic cases of Candida auris in Colombia
Isolation of Candida auris from ear of otherwise healthy patient, Austria, 2018
DOI:10.3201/eid2408.180495
URL
PMID:30016243
[本文引用: 1]

The emerging pathogen Candida auris is isolated mostly from hospitalized patients and often shows multidrug resistance. We report on the isolation of this yeast in Austria from an outpatient's auditory canal. The isolate showed good susceptibility against antifungals except for echinocandins; the patient was treated successfully with topical administration of nystatin.
Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other Candida species
DOI:10.1017/ice.2017.127
URL
PMID:28693657
[本文引用: 2]

Contaminated surfaces have been implicated as a potential route for dissemination of the emerging multidrug-resistant fungal pathogen Candida auris. In laboratory testing, C. auris and other Candida species persisted for 7 days on moist or dry surfaces. Candida species were recovered frequently from the hospital environment, particularly from moist surfaces. Infect Control Hosp Epidemiol 2017;38:1107-1109.
Candida auris: epidemiological situation, laboratory capacity and preparedness in the European Union and European Economic Area*, January 2018 to May 2019
Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism
DOI:10.1016/j.cmi.2015.10.022 URL [本文引用: 2]
Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection- Scotland, 2012-2013
DOI:10.1016/j.cmi.2015.09.018
URL
PMID:26432192
[本文引用: 1]

Bloodstream infections caused by Candida species remain a significant cause of morbidity and mortality in hospitalized patients. Biofilm formation by Candida species is an important virulence factor for disease pathogenesis. A prospective analysis of patients with Candida bloodstream infection (n = 217) in Scotland (2012-2013) was performed to assess the risk factors associated with patient mortality, in particular the impact of biofilm formation. Candida bloodstream isolates (n = 280) and clinical records for 157 patients were collected through 11 different health boards across Scotland. Biofilm formation by clinical isolates was assessed in vitro with standard biomass assays. The role of biofilm phenotype on treatment efficacy was also evaluated in vitro by treating preformed biofilms with fixed concentrations of different classes of antifungal. Available mortality data for 134 patients showed that the 30-day candidaemia case mortality rate was 41%, with predisposing factors including patient age and catheter removal. Multivariate Cox regression survival analysis for 42 patients showed a significantly higher mortality rate for Candida albicans infection than for Candida glabrata infection. Biofilm-forming ability was significantly associated with C. albicans mortality (34 patients). Finally, in vitro antifungal sensitivity testing showed that low biofilm formers and high biofilm formers were differentially affected by azoles and echinocandins, but not by polyenes. This study provides further evidence that the biofilm phenotype represents a significant clinical entity, and that isolates with this phenotype differentially respond to antifungal therapy in vitro. Collectively, these findings show that greater clinical understanding is required with respect to Candida biofilm infections, and the implications of isolate heterogeneity.
Candida biofilms: an update
DOI:10.1128/EC.4.4.633-638.2005 URL PMID:15821123 [本文引用: 1]
Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris
DOI:10.1038/s41426-018-0045-x
URL
PMID:29593275
[本文引用: 2]

Candida auris was first described in 2009, and it has since caused nosocomial outbreaks, invasive infections, and fungaemia across at least 19 countries on five continents. An outbreak of C. auris occurred in a specialized cardiothoracic London hospital between April 2015 and November 2016, which to date has been the largest outbreak in the UK, involving a total of 72 patients. To understand the genetic epidemiology of C. auris infection both within this hospital and within a global context, we sequenced the outbreak isolate genomes using Oxford Nanopore Technologies and Illumina platforms to detect antifungal resistance alleles and reannotate the C. auris genome. Phylogenomic analysis placed the UK outbreak in the India/Pakistan clade, demonstrating an Asian origin; the outbreak showed similar genetic diversity to that of the entire clade, and limited local spatiotemporal clustering was observed. One isolate displayed resistance to both echinocandins and 5-flucytosine; the former was associated with a serine to tyrosine amino acid substitution in the gene FKS1, and the latter was associated with a phenylalanine to isoleucine substitution in the gene FUR1. These mutations add to a growing body of research on multiple antifungal drug targets in this organism. Multiple differential episodic selection of antifungal resistant genotypes has occurred within a genetically heterogenous population across this outbreak, creating a resilient pathogen and making it difficult to define local-scale patterns of transmission and implement outbreak control measures.
First case of Candida auris in Switzerland: discussion about preventive strategies
Candida auris candidaemia in Indian ICUs: analysis of risk factors
DOI:10.1093/jac/dkx034
URL
PMID:28333181
[本文引用: 1]

Objectives: To identify the risk factors associated with Candida auris candidaemia, as this fungus now poses a global threat. Methods: We performed a subgroup analysis of a previously reported study of 27 Indian ICUs. The clinical data of candidaemia cases due to C. auris and other Candida species were compared to determine significant risk factors associated with C. auris infection. Results: Of the 1400 candidaemia cases reported earlier, 74 (5.3%) from 19 of 27 ICUs were due to C. auris . The duration of ICU stay prior to candidaemia diagnosis was significantly longer in patients with C. auris candidaemia (median 25, IQR 12-45 days) compared with the non- auris group (median 15, IQR 9-28, P < 0.001). Based on logistic regression modelling, admission to north Indian ICUs [OR 2.1 (1.2-3.8); P = 0.012], public-sector hospital [OR 2.2 (1.2-3.9); P = 0.006], underlying respiratory illness [OR 2.1 (1.3-3.6); P = 0.002], vascular surgery [OR 2.3 (1.00-5.36); P = 0.048], prior antifungal exposure [OR 2.8 (1.6-4.8); P < 0.001] and low APACHE II score [OR 0.8 (0.8-0.9); P = 0.007] were significantly associated with C. auris candidaemia. The majority (45/51, 88.2%) of the isolates were clonal. A considerable number of isolates were resistant to fluconazole ( n = 43, 58.1%), amphotericin B ( n = 10, 13.5%) and caspofungin ( n = 7, 9.5%). Conclusions: Although C. auris infection has been observed across India, the number of cases is higher in public-sector hospitals in the north of the country. Longer stay in ICU, underlying respiratory illness, vascular surgery, medical intervention and antifungal exposure are the major risk factors for acquiring C. auris infection even among patients showing lower levels of morbidity.
Nosocomial fungemia by Candida auris: first four reported cases in continental Europe
DOI:10.1016/j.riam.2016.11.002 URL [本文引用: 1]
Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris
DOI:10.1128/AAC.00057-19
URL
PMID:30718246
[本文引用: 1]

Candida auris has rapidly emerged as a health care-associated and multidrug-resistant pathogen of global concern. In this work, we examined the relative expression of the four C. auris genes with the highest degree of homology to Candida albicans CDR1 and MDR1 among three triazole-resistant clinical isolates as compared to the triazole-susceptible genome reference clinical isolate. We subsequently utilized a novel Cas9-mediated system for genetic manipulations to delete C. auris CDR1 and MDR1 in both a triazole-resistant clinical isolate and a susceptible reference strain and observed that MICs for all clinically available triazoles decreased as much as 128-fold in the CDR1 deletion strains. The findings of this work reveal for the first time that C. auris CDR1 and MDR1 are more highly expressed among triazole-resistant clinical isolates of C. auris and that the overexpression of CDR1 is a significant contributor to clinical triazole resistance.
Candida auris, an agent of hospital-associated outbreaks: which challenging issues do we need to have in mind?
Differential phospholipase gene expression by Candida albicans in artificial media and cultured human oral epithelium.
Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital
DOI:10.1111/j.1348-0421.2008.00083.x
URL
PMID:19161556
[本文引用: 2]

A single strain of a novel ascomycetous yeast species belonging to the genus Candida was isolated from the external ear canal of an inpatient in a Japanese hospital. Analyses of the 26S rDNA D1/D2 domain, nuclear ribosomal DNA ITS region sequences, and chemotaxonomic studies indicated that this strain represents a new species with a close phylogenetic relationship to Candida ruelliae and Candida haemulonii in the Metschnikowiaceae clade. This strain grew well at 40 degrees C, but showed slow and weak growth at 42 degrees C. The taxonomic description of Candida auris sp. nov. is proposed (type strain JCM15448T= CBS10913T= DSM21092T).
First hospital outbreak of the globally emerging Candida auris in a European hospital
DOI:10.1186/s13756-016-0132-5
URL
PMID:27777756
[本文引用: 2]

BACKGROUND: Candida auris is a globally emerging multidrug resistant fungal pathogen causing nosocomial transmission. We report an ongoing outbreak of C. auris in a London cardio-thoracic center between April 2015 and July 2016. This is the first report of C. auris in Europe and the largest outbreak so far. We describe the identification, investigation and implementation of control measures. METHODS: Data on C. auris case demographics, environmental screening, implementation of infection prevention/control measures, and antifungal susceptibility of patient isolates were prospectively recorded then analysed retrospectively. Speciation of C. auris was performed by MALDI-TOF and typing of outbreak isolates performed by amplified fragment length polymorphism (AFLP). RESULTS: This report describes an ongoing outbreak of 50 C. auris cases over the first 16 month (April 2015 to July 2016) within a single Hospital Trust in London. A total of 44 % (n = 22/50) patients developed possible or proven C. auris infection with a candidaemia rate of 18 % (n = 9/50). Environmental sampling showed persistent presence of the yeast around bed space areas. Implementation of strict infection and prevention control measures included: isolation of cases and their contacts, wearing of personal protective clothing by health care workers, screening of patients on affected wards, skin decontamination with chlorhexidine, environmental cleaning with chorine based reagents and hydrogen peroxide vapour. Genotyping with AFLP demonstrated that C. auris isolates from the same geographic region clustered. CONCLUSION: This ongoing outbreak with genotypically closely related C. auris highlights the importance of appropriate species identification and rapid detection of cases in order to contain hospital acquired transmission.
First reported case of multidrug-resistant Candida auris in Canada
DOI:10.14745/ccdr.v43i78a02
URL
PMID:29770082
[本文引用: 1]

Candida auris is a fungal pathogen that has recently emerged as a global threat to public health. It was first described in Japan in 2009 and has since been reported in 17 countries on five continents. This case report describes the first reported case of multidrug-resistant C. auris in Canada. In May 2017, a 64-year-old individual was evaluated for chronic otitis externa. Past medical history included a recent hospitalization in India for elective oral surgery that was complicated by an odontogenic brain abscess. Upon return to Canada, the individual was admitted to a hospital for neurosurgical drainage of the brain abscess and parenteral antibiotics. Early during hospitalization, the patient was identified as a carrier of carbapenem-resistant Enterobacteriaceae and was placed on contact precautions. Also early during this hospitalization, a chronic otitis media was managed with placement of a typanostomy tube with drainage of clear fluid from the ear, which continued through the admission and after discharge to a post-neurosurgical rehabilitation facility. During outpatient follow-up, swabs of the ear discharge cultured C. auris that was resistant to fluconazole and amphotericin B. There was no clinical response to ototopical antifungal therapy. Surgical evaluation for management of the otomastoiditis is pending. There is a potential for C. auris to cause infection in health care settings. It can persist in hospital environments, has the potential for transmission and can cause invasive disease. It is difficult to identify and is often resistant to antifungal medications. The application of infection prevention and control recommendations can help prevent nosocomial transmission. It is now prudent to consider the risk of C. auris, in addition to the known risk of other antimicrobial resistant organisms, in any traveller who has been hospitalized while outside the country. When identified, contacting local public health can assist in the tracking and management of this emerging disease.
Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR green-based quantitative PCR assay
Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation
DOI:10.1016/j.nmni.2016.07.003
URL
PMID:27617098
[本文引用: 1]

Candida auris is an emerging multidrug resistant yeast that causes nosocomial fungaemia and deep-seated infections. Notably, the emergence of this yeast is alarming as it exhibits resistance to azoles, amphotericin B and caspofungin, which may lead to clinical failure in patients. The multigene phylogeny and amplified fragment length polymorphism typing methods report the C. auris population as clonal. Here, using whole genome sequencing analysis, we decipher for the first time that C. auris strains from four Indian hospitals were highly related, suggesting clonal transmission. Further, all C. auris isolates originated from cases of fungaemia and were resistant to fluconazole (MIC >64 mg/L).
Biofilm-forming capability of highly virulent, multidrug- resistant Candida auris
DOI:10.3201/eid2302.161320
URL
PMID:28098553
[本文引用: 3]

The emerging multidrug-resistant yeast pathogen Candida auris has attracted considerable attention as a source of healthcare-associated infections. We report that this highly virulent yeast has the capacity to form antifungal resistant biofilms sensitive to the disinfectant chlorhexidine in vitro.
The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection
Isolation of Candida auris from cystic fibrosis patient, Greece, April 2019.
Arrival of Candida auris fungus in Singapore: report of the first 3 cases.
Emergence of multidrug-resistant Candida auris in Taiwan
DOI:10.1016/j.ijantimicag.2019.02.011 URL PMID:30822471 [本文引用: 1]
Coevolution of morphology and virulence in Candida species
DOI:10.1128/EC.05085-11
URL
PMID:21764907
[本文引用: 1]

Many of the major human fungal pathogens are known to undergo morphological changes, which in certain cases are associated with virulence. Although there has been an intense research focus on morphology in fungi, very little is known about how morphology evolved in conjunction with a variety of other virulence properties. However, several recent important discoveries, primarily in Candida species, are beginning to shed light on this important area and answer many longstanding questions. In this minireview, we first provide a description of the major fungal morphologies, as well as the roles of morphology and morphology-associated gene expression in virulence. Next, focusing largely on Candida species, we examine the evolutionary relationships among specific morphological forms. Finally, drawing on recent findings, we begin to address the question of how specific morphological changes came to be associated with virulence of Candida species during evolution.
First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China
DOI:10.1038/s41426-018-0131-0
URL
PMID:29992959
[本文引用: 1]

For the first time, we identified 15 cases of Candida auris in Shenyang, China, and then performed a risk factor assessment for these patients compared with 30 control subjects who were hospitalized in the same ward during the same period of time as the infected patients. We found that diarrhea, gastrointestinal decompression, infection, or colonization with other Candida isolates (especially Candida albicans) and tetracycline antibiotics were all risk factors for C. auris infection or colonization. Diarrhea and tetracycline antibiotics were independent risk factors. We suggest clinicians pay special attention to the emergence of multidrug-resistant C. auris infections or colonization.
Phospholipase, proteinase and haemolytic activities of Candida albicans isolated from oral cavities of patients with type 2 diabetes mellitus
DOI:10.1099/jmm.0.47303-0 URL [本文引用: 1]
Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-August 2016
DOI:10.1111/ajt.14121
URL
PMID:28029734
[本文引用: 1]

November 11, 2016/65(44);1234-1237. What is already known about this topic? Candida auris is an emerging pathogenic fungus that has been reported from at least a dozen countries on four continents during 2009-2015. The organism is difficult to identify using traditional biochemical methods, some isolates have been found to be resistant to all three major classes of antifungal medications, and C. auris has caused health care-associated outbreaks. What is added by this report? This is the first description of C. auris cases in the United States. C. auris appears to have emerged in the United States only in the last few years, and U.S. isolates are related to isolates from South America and South Asia. Evidence from U.S. case investigations suggests likely transmission of the organism occurred in health care settings. What are the implications for public health practice? It is important that U.S. laboratories accurately identify C. auris and for health care facilities to implement recommended infection control practices to prevent the spread of C. auris. Local and state health departments and CDC should be notified of possible cases of C. auris and of isolates of C. haemulonii and Candida spp. that cannot be identified after routine testing.
The first two cases of Candida auris in the Netherlands
DOI:10.3390/jof5040091 URL [本文引用: 1]
The first isolate of Candida auris in China: clinical and biological aspects
DOI:10.1038/s41426-018-0095-0
URL
PMID:29777096
[本文引用: 4]

The emerging human fungal pathogen Candida auris has been recognized as a multidrug resistant species and is associated with high mortality. This fungus was first described in Japan in 2009 and has been reported in at least 18 countries on five continents. In this study, we report the first isolate of C. auris from the bronchoalveolar lavage fluid (BALF) of a hospitalized woman in China. Interestingly, this isolate is susceptible to all tested antifungals including amphotericin B, fluconazole, and caspofungin. Copper sulfate (CuSO4) also has a potent inhibitory effect on the growth of this fungus. Under different culture conditions, C. auris exhibits multiple morphological phenotypes including round-to-ovoid, elongated, and pseudohyphal-like forms. High concentrations of sodium chloride induce the formation of a pseudohyphal-like form. We further demonstrate that C. auris is much less virulent than Candida albicans in both mouse systemic and invertebrate Galleria mellonella models.
Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface
DOI:10.1128/JCM.00921-17
URL
PMID:28747370
[本文引用: 3]

The emerging multidrug-resistant pathogenic yeast Candida auris represents a serious threat to global health. Unlike most other Candida species, this organism appears to be commonly transmitted within health care facilities and causes health care-associated outbreaks. To better understand the epidemiology of this emerging pathogen, we investigated the ability of C. auris to persist on plastic surfaces common in health care settings compared with that of Candida parapsilosis, a species known to colonize the skin and plastics. Specifically, we compiled comparative and quantitative data essential to understanding the vehicles of spread and the ability of both species to survive and persist on plastic surfaces under controlled conditions (25 degrees C and 57% relative humidity), such as those found in health care settings. When a test suspension of 10(4) cells was applied and dried on plastic surfaces, C. auris remained viable for at least 14 days and C. parapsilosis for at least 28 days, as measured by CFU. However, survival measured by esterase activity was higher for C. auris than C. parapsilosis throughout the 28-day study. Given the notable length of time Candida species survive and persist outside their host, we developed methods to more effectively culture C. auris from patients and their environment. Using our enrichment protocol, public health laboratories and researchers can now readily isolate C. auris from complex microbial communities (such as patient skin, nasopharynx, and stool) as well as environmental biofilms, in order to better understand and prevent C. auris colonization and transmission.
Analysis of a Candida auris outbreak provides new insights into an emerging pathogen
DOI:10.1128/JCM.02083-19
URL
PMID:31996439
[本文引用: 1]

Candida auris is an emerging human fungal pathogen that is being increasingly linked to outbreaks. It is concerning to health care workers because of its high mortality rate, due primarily to its antifungal resistance. Among the tools being developed to study this yeast are large cohorts of regional isolates, which can be useful for studying epidemiology, antifungal susceptibility patterns, and diagnostic methods. In this issue of the Journal of Clinical Microbiology, Y. Zhu, B. O'Brien, L. Leach, A. Clarke, et al. (J Clin Microbiol 58:e01503-19, 2020, https://doi.org/10.1128/JCM.01503-19) describe the laboratory findings of a collection of isolates from a large outbreak of C. auris obtained from numerous health care facilities in the New York area. Real-time PCR was used as a screening tool with great accuracy, while internal transcribed spacer (ITS) and D1/D2 sequencing were successfully employed for isolate clade assignment. South Asia clade I was identified as the major genotype, while South American clade IV was a minor genotype. Surveillance isolates from patients confirmed axilla/groin and nare colonization; however, results of quantitative analysis of fungal burdens showed that when the nares are colonized, burdens are significantly higher than for axilla/groin colonization. Antifungal susceptibility testing was in agreement with past studies. High levels of fluconazole resistance were detected, while few isolates were resistant to echinocandins. Resistance to multiple antifungals was frequent, and three isolates were recovered that appeared to be pan-resistant. This type of study is yet another useful tool for investigating C. auris, which is becoming an increasingly important human fungal pathogen that should be monitored very closely.
Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch
DOI:10.1038/s41426-018-0187-x
URL
PMID:30482894
[本文引用: 1]

Morphological plasticity has historically been an indicator of increased virulence among fungal pathogens, allowing rapid adaptation to changing environments. Candida auris has been identified as an emerging multidrug-resistant human pathogen of global importance. Since the discovery of this species, it has been thought that C. auris is incapable of filamentous growth. Here, we report the discovery of filamentation and three distinct cell types in C. auris: typical yeast, filamentation-competent (FC) yeast, and filamentous cells. These cell types form a novel phenotypic switching system that contains a heritable (typical yeast-filament) and a nonheritable (FC-filament) switch. Intriguingly, the heritable switch between the typical yeast and the FC/filamentous phenotype is triggered by passage through a mammalian body, whereas the switch between the FC and filamentous phenotype is nonheritable and temperature-dependent. Low temperatures favor the filamentous phenotype, whereas high temperatures promote the FC yeast phenotype. Systemic in vivo and in vitro investigations were used to characterize phenotype-specific variations in global gene expression, secreted aspartyl proteinase (SAP) activity, and changes in virulence, indicating potential for niche-specific adaptations. Taken together, our study not only sheds light on the pathogenesis and biology of C. auris but also provides a novel example of morphological and epigenetic switching in fungi.
Discovery of the gray phenotype and white-gray-opaque tristable phenotypic transitions in Candida dubliniensis
DOI:10.1080/21505594.2015.1135287
URL
PMID:26714067
[本文引用: 1]

Candida dubliniensis is closely related to Candida albicans, a major causative agent of candidiasis, and is primarily associated with oral colonization and infection in human immunodeficiency virus (HIV)-positive patients. Despite the high similarity of genomic and phenotypic features between the 2 species, C. dubliniensis is much less virulent and less prevalent than C. albicans. The ability to change morphological phenotypes is a striking feature of Candida species and is linked to virulence. In this study, we report a novel phenotype, the gray phenotype, in C. dubliniensis. Together with the previously reported white and opaque cell types, the gray phenotype forms a tristable phenotypic switching system in C. dubliniensis that is similar to the white-gray-opaque tristable switching system in C. albicans. Gray cells of C. dubliniensis are similar to their counterparts in C. albicans in terms of several biological aspects including cellular morphology, mating competence, and genetic regulatory mechanisms. However, the gray phenotypes of the 2 species have some distinguishing features. For example, the secreted aspartyl protease (Sap) activity is induced by bovine serum albumin (BSA) in gray cells of C. albicans, but not in gray cells of C. dubliniensis. Taken together, our results demonstrate that the biological features and regulatory mechanisms of white-gray-opaque tristable transitions are largely conserved in the 2 pathogenic Candida species.
The gray phenotype and tristable phenotypic transitions in the human fungal pathogen Candida tropicalis
DOI:10.1016/j.fgb.2016.05.006
URL
PMID:27246518
[本文引用: 1]

Phenotypic plasticity, the ability to switch between different morphological types, plays critical roles in environmental adaptation, leading to infections, and allowing for sexual reproduction in pathogenic Candida species. Candida tropicalis, which is both an emerging human fungal pathogen and an environmental fungus, can switch between two heritable cell types termed white and opaque. In this study, we report the discovery of a novel phenotype in C. tropicalis, named the gray phenotype. Similar to Candida albicans and Candida dubliniensis, white, gray, and opaque cell types of C. tropicalis also form a tristable switching system, where gray cells are relatively small and elongated. In C. tropicalis, gray cells exhibit intermediate levels of mating competency and virulence in a mouse systemic infection model compared to the white and opaque cell types, express a set of cell type-enriched genes, and exhibit both common and species-specific biological features. The key regulators of white-opaque transitions, Wor1 and Efg1, are not required for the gray phenotype. A comparative study of the gray phenotypes in C. tropicalis, C. albicans, and C. dubliniensis provides clues to explain the virulence properties and niche preferences of C. tropicalis.