篮状菌属Talaromyces C.R. Benj.于1955年建立(Benjamin 1955),隶属于真菌界Fungi,子囊菌门Ascomycota,散囊菌纲Eurotiomycetes,散囊菌目Eurotiales,发菌科Trichocomaceae。篮状菌通常产生类似于青霉Penicillium Link无性繁殖结构的帚状枝(penicillus),因此篮状菌最初被当做青霉来研究,但其帚状枝通常为对称双轮生(symmetrical biverticillate),有些种还会产生有性繁殖的子囊果称为裸囊壳(gymnothecium),而青霉的帚状枝通常为单轮生、两轮生、三轮生甚至不规则生,其有性繁殖子囊果为闭囊壳(cleistothecium)。Raper & Thom(1949)在其专著《青霉手册》(《A manual of penicillia》)中,无论有性型和无性型将篮状菌划归在双轮对称组(Penicillium section Biverticillata- Symmetrica Thom)中。Pitt(1979)将只发现无性型的篮状菌物种放在subgenus Biverticillium Pitt中,将产生裸囊壳的种放在Talaromyces中。基于rDNA ITS1-5.8S-ITS2(ITS)、β-微管蛋白基因(β-tubulin gene,BenA)、钙调蛋白基因(calmodulin gene,CaM)和RNA多聚酶II第二大亚基基因(DNA-dependent RNA polymerase II second largest subunit gene,Rpb2)的分子系统学(molecular phylogenetics)研究显示双轮亚属和篮状菌属的种与青霉其他亚属的种分属不同的演化支(clade)(Wang & Zhuang 2007;Houbraken & Samson 2011;Samson et al. 2011)。2012年的《国际藻类、真菌和植物命名法规》(墨尔本法规)[International code of nomenclature for algae,fungi,and plants (Melbourne Code)]认可Talaromyces为上述青霉属双轮亚属和篮状菌属物种的合法属名(McNeill et al. 2012)。
Talaromyces目前共分8个组(section)约175种(Yilmaz et al. 2014;Houbraken et al. 2020;Sun et al. 2020;Wei et al. 2021)。篮状菌组sect. Talaromyces是篮状菌属的最大组,截至本文投稿时全球该组已经报道76个种,我国报道了36个种(Tzean et al. 1994;孔华忠和王龙 2007;Chen et al. 2016;Wang et al. 2016a;Wang et al. 2016b;Wang et al. 2017;Jiang et al. 2018;Su & Niu 2018;陈晗等2021;Houbraken et al. 2020;王龙等 2020;Sun et al. 2020;孙剑秋等2021;单夏男等2021;王龙等2021;Wei et al. 2021)。本研究报道从我国6个省的土壤和纸张样品中分离得到6株篮状菌,经多相分类学鉴定为6个种,即糙孢篮状菌组sect. Trachyspermi的艾斯尤特篮状菌T. assiutensis及sect. Talaromyces的紧密篮状菌T. adpressus、梭形篮状菌T. fusiformis、川芎篮状菌T. cnidii、苹果篮状菌T. malicola和丘陵篮状菌T. tumuli,其中T. cnidii、T. malicola和T. tumuli确定为我国新记录种。
1 材料与方法
1.1 样品采集和分离
样品采自我国浙江、山东、山西、贵州、四川、河北,其中土壤样品取表层土下面的富含腐殖质的土壤约20g置于无菌的塑料袋中封好,纸张样品约5g剪碎后放入无菌的塑料袋中封好。样品分离采用改进的倍比稀释倾倒平皿法(Malloch 1981),即采用0.1%琼脂水溶液(agar water solution)代替无菌水制作样品悬浊液并将倾倒平皿改为涂布平皿(王龙等2020),分离得到的6株篮状菌经鉴定后将每个种的菌株保存于中国普通微生物菌种保藏中心(CGMCC)。
1.2 形态学研究方法
菌落形态研究采用查氏酵母精琼脂(Czapek yeast autolysate agar,CYA)于25℃、37℃、5℃和麦芽精琼脂(5% malt extract agar,MEA)于25℃培养7d后观察、描述和照相,颜色的描述参照Ridgway(1912)的色谱,显微结构研究挑取在MEA 25℃培养7d产生的分生孢子结构做光学显微镜载片观察、照相、描述并鉴定(Pitt 1979;Samson et al. 2010;Yilmaz et al. 2014)。
1.3 PCR扩增和测序
基因组DNA的提取参考Wang & Zhuang(2004)的方法,扩增BenA的引物为bt2a和bt2b(Glass & Donaldson 1995),Rpb2引物为T1和E2(Jiang et al. 2018),ITS的引物为ITS5和ITS4(White et al. 1990)。PCR扩增反应在无菌的0.2mL薄壁平盖Eppendorf管中进行,20µL反应体系含有基因组DNA 1.0µL,正向和反向引物(10µmol/L)各0.5µL,双蒸水8µL,2×PCR扩增缓冲液(0.05U/µL Taq polymerase,4mmol/L MgCl2,0.4mmol/L dNTPs)10µL。PCR程序为94℃预变性3min,然后进行30个循环:94℃变性30s,50℃退火30s,72℃延伸30s,最后72℃延伸5min。PCR产物各取5µL与5µL的100bp DNA ladder用2.0%的琼脂糖凝胶(agarose gel)在80V电压下电泳15min,用0.5g/mL的溴乙锭(ehidium bromide,EB)染色10min后在波长365nm和254nm的紫外灯下观察。显示单一、明亮扩增区段长度条带的PCR扩增产物(BenA约400bp,Rpb2约800bp,ITS约600bp)由睿博兴科生物技术有限公司用ABI3730(Applied Biosystems,Drive Foster City,CA,USA)进行双向直通测序。
1.4 分子系统学分析
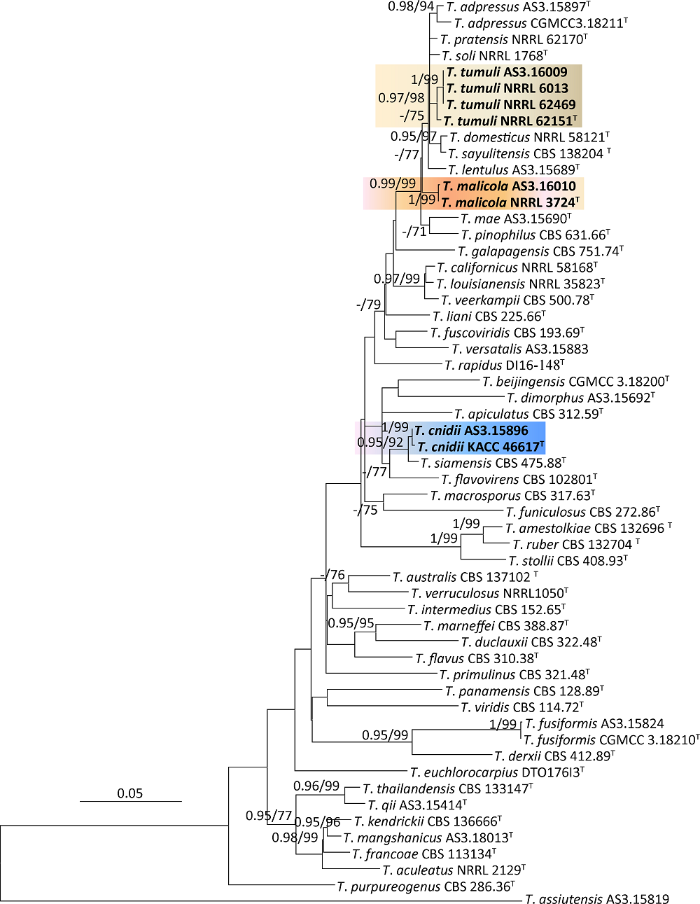
测序得到的原始序列用软件Bioedit 7.0.9(Hall 1999)进行人工校对、编辑,得到准确无误的全区段序列后提交到GenBank。选择篮状菌组48个物种的模式菌株及代表菌株50株和本研究的6个菌株进行分子系统学分析,以T. assiutensis作为外群。这些菌株的BenA、Rpb2、ITS序列用MEGA 6(Tamura et al. 2013)分别进行对位排列(alignment)并编辑修剪后做成序列矩阵,然后用最大似然法(maximum likelihood,ML)分析并采用自展法(bootstrap)进行1 000次重复评估各分支的可靠性,其中空格(gap)选择partial deletion(Hall 2013);这些序列矩阵还采用贝叶斯法(Bayes inference,BI)进行后验概率分析(Posterior probability,PP)(Ronquist et al. 2012)。首先进行BenA、Rpb2和ITS序列的单独分析以检查其谱系一致性(genealogical concordance)(Taylor et al. 2000),然后再连接成BenA-Rpb2-ITS组合序列进行统一分析(图1)。
图1
图1
基于BenA-Rpb2-ITS的ML系统发育树
BI后验概率大于等于0.95和Bootstrap支持率大于等于70%的分支标注在分支节点处,T表示模式菌株,粗体表示新记录种;标尺=0.05每核苷酸替代率
Fig. 1
The ML phylogram inferred from the concatenated BenA-Rpb2-ITS partial sequences.
Posterior probabilities of BI over 0.95 and percentages over 70% derived from 1 000 replicates are indicated at the nodes, T indicates ex-type strains, and the species new to China are indicated in boldface. Bar=0.05 substitutions per nucleotide position.
2 结果与分析
经BenA、Rpb2、ITS分别进行的分子种系学分析显示,这3个遗传标记均得到谱系一致的结果(数据未展示),该3个遗传标记的序列链接后成为BenA-Rpb2-ITS组合序列矩阵共1 524个位点(site),ML最适替代模型为K2+GI。基于该组合序列矩阵的分子系统学分析显示菌株AS3.15896与T. cnidii的模式菌株KACC 46617同在一个分支,Bootstrap支持率为99%,BI后验概率为1;菌株AS3.16010与T. malicola的模式菌株NRRL 3724同在一个分支,Bootstrap支持率为99%,BI后验概率为1;菌株AS3.16009与T. tumuli的模式菌株NRRL 62151等3株菌同在一个分支,Bootstrap支持率为98%,BI后验概率为0.97。依据形态学和分子系统学分析确认这些菌株的鉴定准确无误,参考我国已报道的篮状菌物种,确定这3个种均为我国新记录种(图1-图4)。
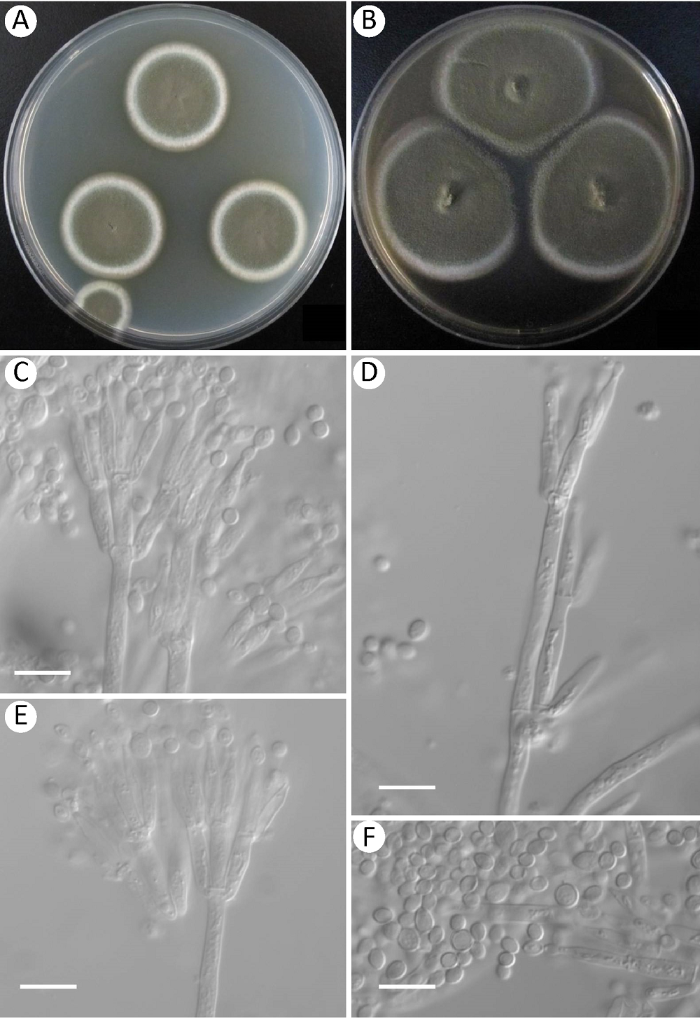
图2
图2
蛇床篮状菌Talaromyces cnidii AS3.15896的形态学性状
A,B:在CYA和MEA上25℃培养7d的菌落;C-E:分生孢子梗;F:分生孢子. 标尺=10μm
Fig. 2
Morphology of Talaromyces cnidii AS3.15896.
A, B: Colonies on CYA and MEA at 25°C in 7d; C-E: Conidiophores; F: Conidia. Scale bars=10μm.
图3
图3
苹果篮状菌Talaromyces malicola AS3.16010的形态学性状
A,B:在CYA和MEA上25℃、7d的菌落;C-E:分生孢子梗;F:分生孢子. 标尺=10μm
Fig. 3
Morphology of Talaromyces malicola AS3.16010.
A, B: Colonies on CYA and MEA at 25°C in 7d; C-E: Conidiophores; F: Conidia. Scale bars=10μm.
图4
图4
丘陵篮状菌Talaromyces tumuli AS3.16009的形态学性状
A,B:在CYA和MEA上25℃、7d的菌落;C-E:分生孢子梗;F:分生孢子. 标尺=10μm
Fig. 4
Morphology of Talaromyces tumuli AS3.16009.
A, B: Colonies on CYA and MEA at 25°C in 7d; C-E: Conidiophores; F: Conidia. Scale bars=10μm.
2.1 蛇床篮状菌
图2Talaromyces cnidii S.H. Yu, T.J. An & H.K. Sang, Journal of Microbiology 51: 707, 2013.
在查氏酵母精琼脂(CYA)上25℃、7d,菌落直径28-30mm,薄,平坦,边缘于培养基内,整齐;质地绒状;分生孢子结构大量,呈橄榄灰绿色Olive-Gray to Dark Olive-Gray(R. Pl. LI);菌丝体呈白色;渗出液无;可溶性色素无;背面中部呈胭脂掌红色Nopal Red(R. Pl. I),外围呈浅橙黄色Light Orange- Yellow(R. Pl. III)。
在麦芽精琼脂(MEA)上25℃、7d,菌落直径42-44mm,薄,平坦,中央稍突起,边缘于培养基表面,整齐;质地绒状;分生孢子结构大量,橄榄灰绿色,近于Olive-Citrine(R. Pl. XVI);菌丝体呈白色;渗出液无;可溶性色素无;背面中央呈红色,其余褐黄色。
在CYA上37℃、7d,正常生长,形成15-17mm的菌落,类似于CYA上25℃。在CYA上5℃、7d,未生长。
分生孢子梗发生于基内菌丝,孢梗茎150-350×3-3.5μm,壁光滑;帚状枝双轮生,排列不紧密,偶尔不规则生;梗基每轮4-6个,排列不紧密,9-12×2.5-3μm;瓶梗披针形,每轮2-6个,10-12×2-3μm;分生孢子椭球形至卵形,3.5-4×2.5-3μm,有些较大,可达5μm,壁光滑。
分布和基物:山西太原纸张(TY7121-11= AS3.15896)。
注:该种生长较快,形成典型绒状菌落,在37℃正常生长;帚状枝双轮生,排列不紧密,偶见不规则生;分生孢子椭球形至卵形,大小不一,壁光滑。
2.2 苹果篮状菌
图3Talaromyces malicola Jurjević & S.W. Peterson, Fungal Biology 123: 756, 2019.
在查氏酵母精琼脂(CYA)上25℃、7d,菌落直径28-30mm,较薄,平坦,边缘于培养基内,流苏状;质地绒状覆盖壳粉色絮状及绳状菌丝体Shell Pink(R. Pl. XXVIII);分生孢子结构稀疏,橄榄灰色Olive Gray(R. Pl. LI);菌丝体在边缘呈白色,近边缘呈浅油黄色Cream Color(R. Pl. XVI);渗出液和可溶性色素无;背面呈浅赭黄色Light Ochraceous Buff(R. Pl. XV)。
在麦芽精琼脂(MEA)上25℃、7d,菌落直径40-41mm,较薄,平坦,边缘于培养内,整齐;质地绒状兼短絮绳状;分生孢子结构大量,豆绿色Pea Green(R. Pl. XLVII);菌丝体呈浅绿黄色Light Green-Yellow(R. Pl. V);渗出液无;可溶性色素无;背面呈殖民黄色Colonial Buff(R. Pl. XXX)。
在CYA上37℃、7d,菌落直径9-10mm,类似于CYA上25℃。
在CYA上5℃、7d,未生长。
分生孢子梗发生于表面菌丝和气生菌丝,孢梗茎50-150×3-3.5μm,孢子壁光滑;帚状枝主要双轮生,偶尔单轮生;梗基每轮6-10个,8-12×3-4μm;瓶梗披针形,每轮6-8个,9-12×2.5-3μm;分生孢子球形至近球形,3-4×2.5-3μm,壁光滑至稍粗糙。
主要特征:生长适中,菌丝体呈白色兼淡粉色,分生孢子灰绿色,在MEA上的菌落形态类似青霉,在37℃正常生长;帚状枝主要双轮生,偶尔单轮生,分生孢子球形至近球形,壁光滑至稍粗糙。
分布和基物:四川成都彭州县土壤(PZ3-2=AS3.16010)。
2.3 丘陵篮状菌
图4Talaromyces tumuli Jurjević & S. W. Peterson, Fungal Biology 123: 758, 2019.
在查氏酵母精琼脂(CYA)上25℃培养7d,菌落直径23-28mm,较薄,表面具适量辐射状沟纹,边缘于培养基内部,整齐;质地绒状兼短絮绳状;分生孢子结构大量,灰绿色,近于豆绿色Pea Green to Gnaphalium Green(R. Pl. XLVII);菌丝体呈白色;渗出液无或少量,无色;可溶性色素无;背面呈赭黄色Yellow Ocher(R. Pl. XV)。
在麦芽精琼脂(MEA)上25℃培养7d,菌落直径48-50mm,较薄,平坦,边缘于培养内部,规则;质地绒状兼绳状;分生孢子结构大量,灰绿色,近于豆绿色Pea Green(R. Pl. XLVII);菌丝体呈白色;无渗出液;无可溶性色素;背面呈褐黄色,近于Colonial Buff(R. Pl. XXX)。
在CYA上37℃、7d,菌落直径23-25mm,类似于CYA上25℃。
在CYA上5℃、7d,未生长。
分生孢子梗发生于表面菌丝和绳状菌丝,孢梗茎50-200×3-4μm,孢子壁光滑;帚状枝主要双轮生,偶尔单轮生和不规则生;梗基每轮8-12个,8-12×3-4μm;瓶梗披针形,每轮6-10个,9-10×2.5-3μm;分生孢子椭球形至柠檬形,3-4×2.5-3μm,壁光滑至稍粗糙。
主要特征:生长适中,菌丝体呈白色,分生孢子灰绿色,在MEA上的菌落形态类似青霉,在37℃正常生长;帚状枝主要双轮生,偶尔单轮生,分生孢子椭球形至柠檬形,壁光滑至稍粗糙。
分布和基物:河北蠡县土壤(LX2-4=AS 3.16009)。
3 讨论
Talaromyces cnidii在美国、韩国、泰国均有发现(Sang et al. 2013;Visagie et al. 2014;Guevara-Suarez et al. 2017),其中美国的菌株分离自支气管肺泡灌洗液(bronchoalveolar lavage,BAL),泰国的6个菌株分离自室内尘土,模式菌株KACC 46617分离自韩国草药日本川芎Cnidium officinale(蛇床属)干燥后的根,本研究报道的菌株AS3.15896分离自纸张。该种可能广泛分布于人类活动场所,而且能在37℃正常生长,因此有可能对人类健康造成威胁。我国的菌株AS3.15896在菌落形态上与模式菌株几乎完全相同,其帚状枝及各个部分的形态和尺度与模式菌株也非常接近,但其产生一些不规则帚状枝,而模式菌株只产生双轮生帚状枝。该菌株与模式菌株在BenA、Rpb2和ITS序列上分别各有1个碱基的差异。BenA-Rpb2-ITS组合序列的分子种系学分析显示我国菌株与模式菌株同在一个分支且支持率非常显著(图1)。
Talaromyces malicola此前只发现于意大利,而且GenBank只记录了其模式菌株NRRL 3724的序列。我国菌株AS3.16010与模式菌株在CYA菌落形态上非常相似,均形成絮状和绳状脏粉色及牙黄色菌丝体,分生孢子稀疏,呈灰色。但在MEA上我国菌株形成绒状菌落且产生大量分生孢子,而模式菌株形成絮状兼绳状菌落,只产生适量分生孢子。它们的显微结构几乎完全相同,其分生孢子梗均发生于表面菌丝和气生菌丝,帚状枝为双轮生兼单轮生,分生孢子均为球形至近球形,壁光滑至稍粗糙(Peterson & Jurjević 2019)。该菌株与模式菌株在BenA序列上只有2个碱基的差异,在Rpb2和ITS序列上完全相同。组合序列的分子种系学分析显示该菌株与模式菌株同在一个分支且具有显著的支持率(图1)。
Talaromyces tumuli此前发现于美国和南非,GenBank只记录了6株菌的序列。我国的菌株AS 3.16009与模式菌株NRRL 62151在CYA和MEA的菌落形态上很相似,这两株菌均产生絮状兼绳状菌落及大量灰绿色分生孢子,只是我国菌株在MEA上比模式菌株生长较快(48-50mm vs. 36-42mm)。该菌株在显微结构上与模式菌株几乎完全相同,均产生双轮生和单轮生及不规则生帚状枝,排列不紧密,瓶梗安瓿形,分生孢子椭球形至柠檬形,壁光滑至稍粗糙(Peterson & Jurjević 2019)。该菌株与模式菌株在BenA序列上有5个碱基的差异,但与另外2个菌株,即NRRL 6013和NRRL 62469完全相同,在Rpb2序列上与模式菌株及NRRL 6013和NRRL 62469完全相同,在ITS序列上与模式菌株只有1个核苷酸的差异,而与另外2个菌株完全相同。组合序列的分子系统学分析也显示该菌株与T. tumuli的模式菌株及另外2个菌株同在一个分支且具有显著的支持率(图1)。
参考文献
Ascocarps of Aspergillus and Penicillium
DOI:10.2307/3755578 URL [本文引用: 1]
New Talaromyces species from indoor environments in China
DOI:10.1016/j.simyco.2016.11.003
PMID:28070136
[本文引用: 1]

contains both asexual and sexually reproducing species. This genus is divided in seven sections and currently has 105 accepted species. In this study we investigated the isolates that were obtained during a study of indoor air collected in Beijing, China. These indoor strains are resolved in four sections, seven of them are identified as,, and according to sequences, while 14 isolates have divergent sequences and are described here as nine new species. The new species are placed in four sections, namely sections,, and. They are described based on sequence data (ITS,, and ) in combination with phenotypic and extrolite characters. Morphological descriptions and notes for distinguishing similar species are provided for each new species. The recently described is synonymised with based on presented phylogenetic results.
Current taxonomy of Talaromyces and three new Chinese records
Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes
DOI:10.1128/aem.61.4.1323-1330.1995
PMID:7747954
[本文引用: 1]

We constructed nine sets of oligonucleotide primers on the basis of the results of DNA hybridization of cloned genes from Neurospora crassa and Aspergillus nidulans to the genomes of select filamentous ascomycetes and deuteromycetes (with filamentous ascomycete affiliations). Nine sets of primers were designed to amplify segments of DNA that span one or more introns in conserved genes. PCR DNA amplification with the nine primer sets with genomic DNA from ascomycetes, deuteromycetes, basidiomycetes, and plants revealed that five of the primer sets amplified a product only from DNA of the filamentous ascomycetes and deuteromycetes. The five primer sets were constructed from the N. crassa genes for histone 3, histone 4, beta-tubulin, and the plasma membrane ATPase. With these five primer sets, polymorphisms were observed in both the size of and restriction enzyme sites in the amplified products from the filamentous ascomycetes. The primer sets described here may provide useful tools for phylogenetic studies and genome analyses in filamentous ascomycetes and deuteromycetes (with ascomycete affiliations), as well as for the rapid differentiation of fungal species by PCR.
Four new species of Talaromyces from clinical sources
Building phylogenetic trees from molecular data with MEGA
DOI:10.1093/molbev/mst012 URL [本文引用: 1]
BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT
Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species
DOI:10.1016/j.simyco.2020.05.002
PMID:32855739
[本文引用: 2]

The is a relatively large order of with members frequently having positive and negative impact on human activities. Species within this order gain attention from various research fields such as food, indoor and medical mycology and biotechnology. In this article we give an overview of families and genera present in the and introduce an updated subgeneric, sectional and series classification for and. Finally, a comprehensive list of accepted species in the is given. The classification of the at family and genus level is traditionally based on phenotypic characters, and this classification has since been challenged using sequence-based approaches. Here, we re-evaluated the relationships between families and genera of the using a nine-gene sequence dataset. Based on this analysis, the new family is introduced and four known families are accepted:,, and. The includes 28 genera: 15 genera are accommodated in the (,,,,,,,,,,,,, and ), eight in the (,,,,,,, ), two in the (, ) and one in the (). The classification of the was not part of this study, but according to literature two genera are present in this family ( and ). The use of an infrageneric classification system has a long tradition in and. Most recent taxonomic studies focused on the sectional level, resulting in a well-established sectional classification in these genera. In contrast, a series classification in and is often outdated or lacking, but is still relevant,, the allocation of a species to a series can be highly predictive in what functional characters the species might have and might be useful when using a phenotype-based identification. The majority of the series in and are invalidly described and here we introduce a new series classification. Using a phylogenetic approach, often supported by phenotypic, physiologic and/or extrolite data, is subdivided in six subgenera, 27 sections (five new) and 75 series (73 new, one new combination), and in two subgenera, 32 sections (seven new) and 89 series (57 new, six new combinations). Correct identification of species belonging to the is difficult, but crucial, as the species name is the linking pin to information. Lists of accepted species are a helpful aid for researchers to obtain a correct identification using the current taxonomic schemes. In the most recent list from 2014, 339, 354 and 88 species were accepted. These numbers increased significantly, and the current list includes 446 (32 % increase), 483 (36 % increase) and 171 (94 % increase) species, showing the large diversity and high interest in these genera. We expanded this list with all genera and species belonging to the (except those belonging to ). The list includes 1 187 species, distributed over 27 genera, and contains MycoBank numbers, collection numbers of type and ex-type cultures, subgenus, section and series classification data, information on the mode of reproduction, and GenBank accession numbers of ITS, beta-tubulin (), calmodulin () and RNA polymerase II second largest subunit () gene sequences.© 2020 Westerdijk Fungal Biodiversity Institute. Production and hosting by ELSEVIER B.V.
Phylogeny of Penicillium and the segregation of Trichocomaceae into three families
DOI:10.3114/sim.2011.70.01
PMID:22308045
[本文引用: 1]

Species of Trichocomaceae occur commonly and are important to both industry and medicine. They are associated with food spoilage and mycotoxin production and can occur in the indoor environment, causing health hazards by the formation of β-glucans, mycotoxins and surface proteins. Some species are opportunistic pathogens, while others are exploited in biotechnology for the production of enzymes, antibiotics and other products. Penicillium belongs phylogenetically to Trichocomaceae and more than 250 species are currently accepted in this genus. In this study, we investigated the relationship of Penicillium to other genera of Trichocomaceae and studied in detail the phylogeny of the genus itself. In order to study these relationships, partial RPB1, RPB2 (RNA polymerase II genes), Tsr1 (putative ribosome biogenesis protein) and Cct8 (putative chaperonin complex component TCP-1) gene sequences were obtained. The Trichocomaceae are divided in three separate families: Aspergillaceae, Thermoascaceae and Trichocomaceae. The Aspergillaceae are characterised by the formation flask-shaped or cylindrical phialides, asci produced inside cleistothecia or surrounded by Hülle cells and mainly ascospores with a furrow or slit, while the Trichocomaceae are defined by the formation of lanceolate phialides, asci borne within a tuft or layer of loose hyphae and ascospores lacking a slit. Thermoascus and Paecilomyces, both members of Thermoascaceae, also form ascospores lacking a furrow or slit, but are differentiated from Trichocomaceae by the production of asci from croziers and their thermotolerant or thermophilic nature. Phylogenetic analysis shows that Penicillium is polyphyletic. The genus is re-defined and a monophyletic genus for both anamorphs and teleomorphs is created (Penicillium sensu stricto). The genera Thysanophora, Eupenicillium, Chromocleista, Hemicarpenteles and Torulomyces belong in Penicilliums. str. and new combinations for the species belonging to these genera are proposed. Analysis of Penicillium below genus rank revealed the presence of 25 clades. A new classification system including both anamorph and teleomorph species is proposed and these 25 clades are treated here as sections. An overview of species belonging to each section is presented.New sections, all in Penicillium: sect. Sclerotiora Houbraken & Samson, sect. Charlesia Houbraken & Samson, sect. Thysanophora Houbraken & Samson,sect. Ochrosalmonea Houbraken & Samson, sect. Cinnamopurpurea Houbraken & Samson, Fracta Houbraken & Samson, sect. Stolkia Houbraken & Samson, sect. Gracilenta Houbraken & Samson, sect. Citrina Houbraken & Samson, sect. Turbata Houbraken & Samson, sect. Paradoxa Houbraken & Samson, sect. Canescentia Houbraken & Samson. New combinations:Penicillium asymmetricum (Subramanian & Sudha) Houbraken & Samson, P. bovifimosum (Tuthill & Frisvad) Houbraken & Samson, P. glaucoalbidum (Desmazières) Houbraken & Samson, P. laeve (K. Ando & Manoch) Houbraken & Samson, P. longisporum (Kendrick) Houbraken & Samson, P. malachiteum (Yaguchi & Udagawa) Houbraken & Samson, P. ovatum (K. Ando & Nawawi) Houbraken & Samson, P. parviverrucosum (K. Ando & Pitt) Houbraken & Samson, P. saturniforme (Wang & Zhuang) Houbraken & Samson, P. taiwanense (Matsushima) Houbraken & Samson. New names:Penicillium coniferophilum Houbraken & Samson, P. hennebertii Houbraken & Samson, P. melanostipe Houbraken & Samson, P. porphyreum Houbraken & Samson.
Three new species of Talaromyces sect. Talaromyces discovered from soil in China
DOI:10.1038/s41598-018-23370-x URL [本文引用: 2]
International Code of Nomenclature for algae, fungi, and plants, Melbourne Code, Regnum Vegetabile 154
The Talaromyces pinophilus species complex
DOI:S1878-6146(18)30307-6
PMID:31542192
[本文引用: 2]

A sample of isolates from Talaromyces pinophilus (55 isolates) and closely related species (76 isolates) was sequenced at four loci, the data were analyzed using maximum likelihood analysis and the GCPSR. The isolates were subjected to growth studies on the recommended media for description of Talaromyces species. On the basis of the combined data, five new species were segregated out of T. pinophilus and placed in newly described species. The T. pinophilus species complex contains ten species. The three other new species, Talaromyces argentinensis, T. californicus and T. louisianensis were not a part of the T. pinophilus species complex but occurred in Talaromyces sect. Talaromyces. T. argentinensis produces a teleomorphic state and is phylogenetically and morphologically distinct from other Talaromyces species.Published by Elsevier Ltd.
Color standards and color nomenclature. Published by the author
MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space
DOI:10.1093/sysbio/sys029
PMID:22357727
[本文引用: 1]

Since its introduction in 2001, MrBayes has grown in popularity as a software package for Bayesian phylogenetic inference using Markov chain Monte Carlo (MCMC) methods. With this note, we announce the release of version 3.2, a major upgrade to the latest official release presented in 2003. The new version provides convergence diagnostics and allows multiple analyses to be run in parallel with convergence progress monitored on the fly. The introduction of new proposals and automatic optimization of tuning parameters has improved convergence for many problems. The new version also sports significantly faster likelihood calculations through streaming single-instruction-multiple-data extensions (SSE) and support of the BEAGLE library, allowing likelihood calculations to be delegated to graphics processing units (GPUs) on compatible hardware. Speedup factors range from around 2 with SSE code to more than 50 with BEAGLE for codon problems. Checkpointing across all models allows long runs to be completed even when an analysis is prematurely terminated. New models include relaxed clocks, dating, model averaging across time-reversible substitution models, and support for hard, negative, and partial (backbone) tree constraints. Inference of species trees from gene trees is supported by full incorporation of the Bayesian estimation of species trees (BEST) algorithms. Marginal model likelihoods for Bayes factor tests can be estimated accurately across the entire model space using the stepping stone method. The new version provides more output options than previously, including samples of ancestral states, site rates, site d(N)/d(S) rations, branch rates, and node dates. A wide range of statistics on tree parameters can also be output for visualization in FigTree and compatible software.
Food and indoor fungi (2nd ed.) CBS-KNAW Fungal Biodiversity Centre, Utrecht, the
Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium
DOI:10.3114/sim.2011.70.04
PMID:22308048
[本文引用: 3]

The taxonomic history of anamorphic species attributed to Penicillium subgenus Biverticillium is reviewed, along with evidence supporting their relationship with teleomorphic species classified in Talaromyces. To supplement previous conclusions based on ITS, SSU and/or LSU sequencing that Talaromyces and subgenus Biverticillium comprise a monophyletic group that is distinct from Penicillium at the generic level, the phylogenetic relationships of these two groups with other genera of Trichocomaceae was further studied by sequencing a part of the RPB1 (RNA polymerase II largest subunit) gene. Talaromyces species and most species of Penicillium subgenus Biverticilliumsensu Pitt reside in a monophyletic clade distant from species of other subgenera of Penicillium. For detailed phylogenetic analysis of species relationships, the ITS region (incl. 5.8S nrDNA) was sequenced for the available type strains and/or representative isolates of Talaromyces and related biverticillate anamorphic species. Extrolite profiles were compiled for all type strains and many supplementary cultures. All evidence supports our conclusions that Penicillium subgenus Biverticillium is distinct from other subgenera in Penicillium and should be taxonomically unified with the Talaromyces species that reside in the same clade. Following the concepts of nomenclatural priority and single name nomenclature, we transfer all accepted species of Penicillium subgenus Biverticillium to Talaromyces. A holomorphic generic diagnosis for the expanded concept of Talaromyces, including teleomorph and anamorph characters, is provided. A list of accepted Talaromyces names and newly combined Penicillium names is given. Species of biotechnological and medical importance, such as P. funiculosum and P. marneffei, are now combined in Talaromyces. Excluded species and taxa that need further taxonomic study are discussed. An appendix lists other generic names, usually considered synonyms of Penicillium sensu lato that were considered prior to our adoption of the name Talaromyces.Taxonomic novelties:New species - Talaromyces apiculatus Samson, Yilmaz & Frisvad, sp. nov. New combinationsand names - Talaromyces aculeatus (Raper & Fennell) Samson, Yilmaz, Frisvad & Seifert, T. albobiverticillius (H.-M. Hsieh, Y.-M. Ju & S.-Y. Hsieh) Samson, Yilmaz, Frisvad & Seifert, T. allahabadensis (B.S. Mehrotra & D. Kumar) Samson, Yilmaz & Frisvad, T. aurantiacus (J.H. Mill., Giddens & A.A. Foster) Samson, Yilmaz, & Frisvad, T. boninensis (Yaguchi & Udagawa) Samson, Yilmaz, & Frisvad, T. brunneus (Udagawa) Samson, Yilmaz & Frisvad, T. calidicanius (J.L. Chen) Samson, Yilmaz & Frisvad, T. cecidicola (Seifert, Hoekstra & Frisvad) Samson, Yilmaz, Frisvad & Seifert, T. coalescens (Quintan.) Samson, Yilmaz & Frisvad, T. dendriticus (Pitt) Samson, Yilmaz, Frisvad & Seifert, T. diversus (Raper & Fennell) Samson, Yilmaz & Frisvad, T. duclauxii (Delacr.) Samson, Yilmaz, Frisvad & Seifert, T. echinosporus (Nehira) Samson, Yilmaz & Frisvad, comb. nov. T. erythromellis (A.D. Hocking) Samson, Yilmaz, Frisvad & Seifert, T. funiculosus (Thom) Samson, Yilmaz, Frisvad & Seifert, T. islandicus (Sopp) Samson, Yilmaz, Frisvad & Seifert, T. loliensis (Pitt) Samson, Yilmaz & Frisvad, T. marneffei (Segretain, Capponi & Sureau) Samson, Yilmaz, Frisvad & Seifert, T. minioluteus (Dierckx) Samson, Yilmaz, Frisvad & Seifert, T. palmae (Samson, Stolk & Frisvad) Samson, Yilmaz, Frisvad & Seifert, T. panamensis (Samson, Stolk & Frisvad) Samson, Yilmaz, Frisvad & Seifert, T. paucisporus (Yaguchi, Someya & Udagawa) Samson & Houbraken T. phialosporus (Udagawa) Samson, Yilmaz & Frisvad, T. piceus (Raper & Fennell) Samson, Yilmaz, Frisvad & Seifert, T. pinophilus (Hedgcock) Samson, Yilmaz, Frisvad & Seifert, T. pittii (Quintan.) Samson, Yilmaz, Frisvad & Seifert, T. primulinus (Pitt) Samson, Yilmaz & Frisvad, T. proteolyticus (Kamyschko) Samson, Yilmaz & Frisvad, T. pseudostromaticus (Hodges, G.M. Warner, Rogerson) Samson, Yilmaz, Frisvad & Seifert, T. purpurogenus (Stoll) Samson, Yilmaz, Frisvad & Seifert, T. rademirici (Quintan.) Samson, Yilmaz & Frisvad, T. radicus (A.D. Hocking & Whitelaw) Samson, Yilmaz, Frisvad & Seifert, T. ramulosus (Visagie & K. Jacobs) Samson, Yilmaz, Frisvad & Seifert, T. rubicundus (J.H. Mill., Giddens & A.A. Foster) Samson, Yilmaz, Frisvad & Seifert, T. rugulosus (Thom) Samson, Yilmaz, Frisvad & Seifert, T. sabulosus (Pitt & A.D. Hocking) Samson, Yilmaz & Frisvad, T. siamensis (Manoch & C. Ramírez) Samson, Yilmaz & Frisvad, T. sublevisporus (Yaguchi & Udagawa) Samson, Yilmaz & Frisvad, T. variabilis (Sopp) Samson, Yilmaz, Frisvad & Seifert, T. varians (G. Sm.) Samson, Yilmaz & Frisvad, T. verruculosus (Peyronel) Samson, Yilmaz, Frisvad & Seifert, T. viridulus Samson, Yilmaz & Frisvad.
Two novel Talaromyces species isolated from medicinal crops in Korea
DOI:10.1007/s12275-013-3361-9 URL [本文引用: 1]
Four species of Talaromyces new to China
Multilocus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species, T. cucurbitiradicus and T. endophyticus
DOI:10.1080/00275514.2018.1432221 URL [本文引用: 1]
New section and species in Talaromyces
DOI:10.3897/mycokeys.68.52092 URL
The importance of Talaromyces and its taxonomy
MEGA 6: molecular evolutionary genetics analysis version 6.0
Phylogenetic species recognition and species concepts in fungi
The operational species concept, i.e., the one used to recognize species, is contrasted to the theoretical species concept. A phylogenetic approach to recognize fungal species based on concordance of multiple gene genealogies is compared to those based on morphology and reproductive behavior. Examples where Phylogenetic Species Recognition has been applied to fungi are reviewed and concerns regarding Phylogenetic Species Recognition are discussed.
Penicillium and related teleomorphs from Taiwan
Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world
DOI:10.1016/j.simyco.2014.07.002
PMID:25492981
[本文引用: 1]

As part of a worldwide survey of the indoor mycobiota, dust was collected from nine countries. Analyses of dust samples included the culture-dependent dilution-to-extinction method and the culture-independent 454-pyrosequencing. Of the 7 904 isolates, 2 717 isolates were identified as belonging to Aspergillus, Penicillium and Talaromyces. The aim of this study was to identify isolates to species level and describe the new species found. Secondly, we wanted to create a reliable reference sequence database to be used for next-generation sequencing projects. Isolates represented 59 Aspergillus species, including eight undescribed species, 49 Penicillium species of which seven were undescribed and 18 Talaromyces species including three described here as new. In total, 568 ITS barcodes were generated, and 391 beta-tubulin and 507 calmodulin sequences, which serve as alternative identification markers.
Two new records of Talaromyces section Islandici species from China
Talaromyces cecidicola and T. pseudostromaticus, two species of Talaromyces section Purpurei new to China
Designing primer sets for amplification of partial calmodulin genes from penicillia
Phylogenetic analyses of penicillia based on partial calmodulin gene sequences
DOI:10.1016/j.biosystems.2006.04.008 URL [本文引用: 1]
Talaromyces neofusisporus and T. qii, two new species of section Talaromyces isolated from plant leaves in Tibet, China
DOI:10.1038/srep18622 URL [本文引用: 1]
Talaromyces heiheensis and T. mangshanicus, two new species from China
DOI:10.1007/s11557-016-1251-3 URL [本文引用: 1]
A new species of Talaromyces, Trichocomaceae, from the Xisha Islands, Hainan, China
DOI:10.11646/phytotaxa.267.3 URL [本文引用: 1]
Four new species of Talaromyces sect. Talaromyces discovered in China
DOI:10.1080/00275514.2020.1853457 URL [本文引用: 2]
Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics
Polyphasic taxonomy of the genus Talaromyces
DOI:10.1016/j.simyco.2014.08.001
PMID:25492983
[本文引用: 2]

The genus Talaromyces was described by Benjamin in 1955 as a sexual state of Penicillium that produces soft walled ascomata covered with interwoven hyphae. Phylogenetic information revealed that Penicillium subgenus Biverticillium and Talaromyces form a monophyletic clade distinct from the other Penicillium subgenera. Subsequently, in combination with the recent adoption of the one fungus one name concept, Penicillium subgenus Biverticillium was transferred to Talaromyces. At the time, the new combinations were made based only on phylogenetic information. As such, the aim of this study was to provide a monograph on Talaromyces applying a polyphasic species concept, including morphological, molecular and physiological characters. Based on an ITS, BenA and RPB2 multigene phylogeny, we propose a new sectional classification for the genus, placing the 88 accepted species into seven sections, named sections Bacillispori, Helici, Islandici, Purpurei, Subinflati, Talaromyces and Trachyspermi. We provide morphological descriptions for each of these species, as well as notes on their identification using morphology and DNA sequences. For molecular identification, BenA is proposed as a secondary molecular marker to the accepted ITS barcode for fungi.
虫瘿篮状菌和伪子座篮状菌——篮状菌属紫篮状菌组的两个我国新记录种