香菇Lentinula edodes (Berk.) Pegler是我国食用菌产业中最重要的栽培品种之一,其年产量居于全国第一,基于原生质体单核化进行杂交育种,获得新的性状,产生新的品种,已经成为香菇种质资源创新的重要路径(何志勇等 2000;宋莹等 2016, 2017),同时香菇单核体是一种杂交配子可回溯材料,可以应用于分子标记开发和品种保护等方面(李安政 2006;徐年生等 2012;鲍大鹏 2019)。在试验中同样发现了香菇单核体分离过程中的生长差异和偏分离现象(程水明 2005;程爽爽等 2019;侯娣等 2023;姜珊等 2023),并将此现象定义为强势核和弱势核(姜珊等 2023),但是产生这种生长差异现象的原因至今还没有阐述清楚。研究产生这种差异的分子机制是了解香菇双核体生理学特征、遗传学机制和调控机制的重要的应用性基础研究,可为解决食用真菌菌种培育、种性稳定等方面问题提供理论指导。
1 材料与方法
1.1 供试菌株来源
1.2 培养基制备
本研究使用2种培养基:PDA和添加了2%木屑的PDA,培养基配置方法和木屑处理方法参照侯娣等(2023)的报道。
1.3 香菇菌丝培养方法及数据测量
超净工作台紫外线灭菌30 min后,在香菇单核体菌丝边缘打孔,使用接种针分别接种于PDA培养基和添加2.0%木屑PDA培养基的中间。每个处理设置3个平行样,在25 ℃恒温培养箱避光培养。本研究采取十字交叉法测量菌丝的生长(聂建军等 2020),即平板接种后先对平板进行十字划线,待菌丝萌发,在其生长尖端标记最长半径r1,待双核和单核体菌丝生长3 d、6 d时,在菌丝最外围做标记r2,然后用游标卡尺测量r1与r2之间的距离(mm),再除以菌丝培养天数(d)即得菌丝平均生长速度。所获得的实验数据使用SPSS Statistics 26统计分析软件进行显著性分析(P<0.05)。
1.4 转录组测序样品准备、测序以及数据分析
Y0040-1和Y0040-3这一对异核体在2种培养基上培养9 d开始收集菌丝,每个试验设计设置3个重复,组名分别为CK1、WP1、CK3和WP3。从样品中提取total RNA,利用Nanodrop2000对所提RNA的浓度和纯度进行检测,琼脂糖凝胶电泳检测RNA完整性,RNA 片段长度采用 Agilent 2100检测,样品合格后,文库构建以及高通量测序在上海美吉生物医药科技有限公司完成。原始数据质控以及mapping信息请参照侯娣等(2023)的报道。使用RSEM软件(
2 结果与分析
2.1 生长速度的比较分析
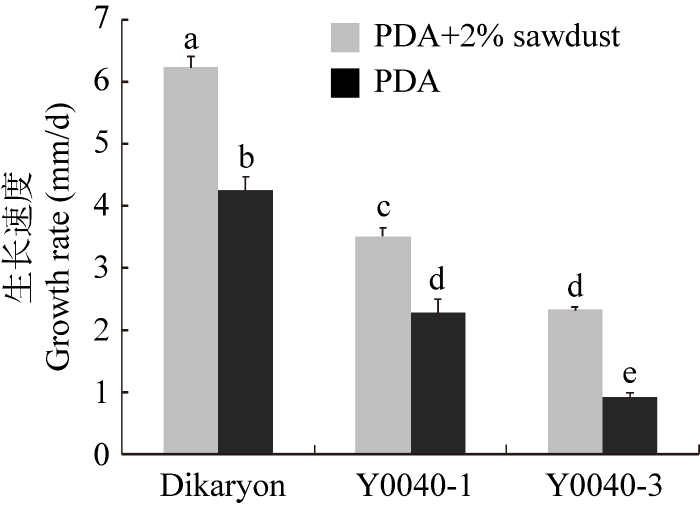
本研究发现从同一出发菌株制备的一对亲和单核体菌丝的生长速度差异明显,其中一个单核体Y0040-1在PDA和木屑-PDA培养基上菌丝增长率明显高于另一单核体Y0040-3,而双核菌丝的生长速度高于单核菌丝(图1)。
图1
图1
双核体、单核体在PDA培养基与2%木屑粉PDA培养基上菌丝生长对比图
不同小写字母表示P<0.05 水平有显著差异
Fig. 1
Comparison of mycelial growth between monokaryon strains Y0040-1 and Y0040-3 on PDA and PDA + 2% sawdust.
Different lowercase letters represent significant level at P<0.05.
2.2 单核转录组测序分析
2.2.1 转录组差异表达基因分析
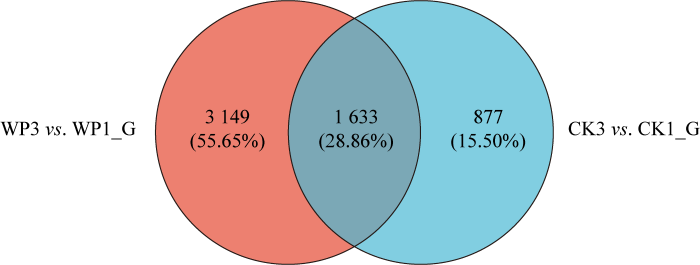
将2个差异核体进行转录组测序和分析,根据DEG的筛选标准|log2FC|>1且qvalue<0.005,2个数据组CK3 vs. CK1、WP3 vs. WP1中共鉴定出5 659个差异基因,分组比较进行韦恩图(Venn)分析发现,CK3 vs. CK1、WP3 vs. WP1差异基因数量分别为2 510个和4 782个,WP3 vs. WP1显著性差异表达的基因数目最多。在这两组中分别有877个、3 149个特有差异表达基因,WP3 vs. WP1组特有差异表达基因数量最高(图2)。
图2
2.2.2 单核体差异表达
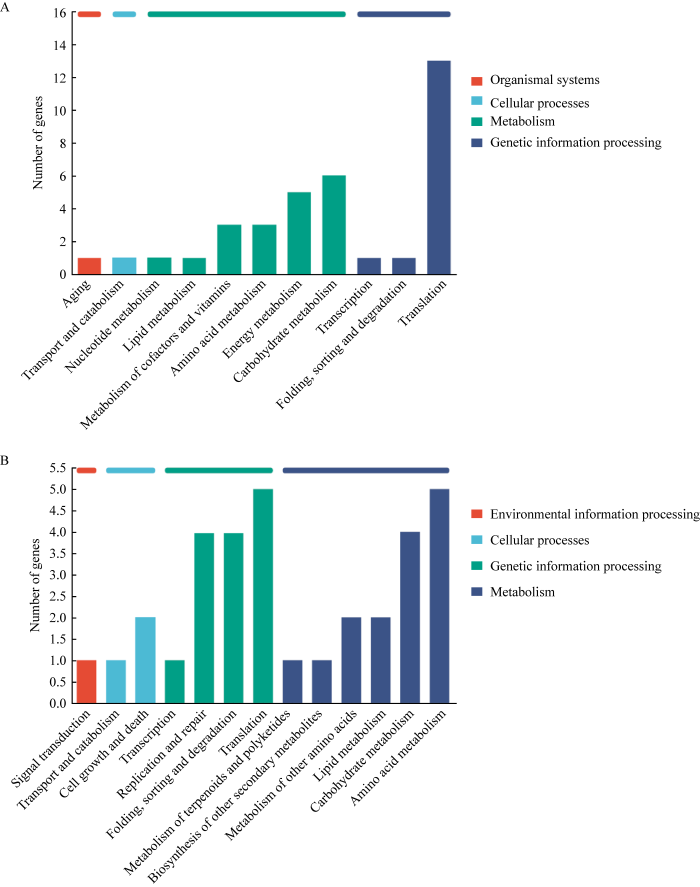
通过Venn分析发现1 633个基因发生了共同的变化,这些共同变化的基因可能是导致两者性状差异的共同表达通路,这些基因中共同上调的基因有155个,共同下调的基因有136个。进一步对这些基因进行KEGG注释分析,发现共同上调基因在氨基酸代谢、能量代谢和碳水化合物代谢等过程中得到富集;而下调的基因主要参与氨基酸代谢、碳水化合物代谢、脂类代谢等过程,可见两者在氨基酸代谢和碳水化合物代谢都得到了富集(图3),而碳水化合物代谢与Cazymes家族基因密切相关。
图3
图3
在比较组CK3 vs. CK1和WP3 vs. WP1共同调节基因的KEGG富集
A:上调基因. B:下调基因
Fig. 3
KEGG enrichment of co-regulated genes in CK3 vs. CK1 and WP3 vs. WP1.
A: Co-upregulated genes. B: Co-downregulated genes.
2.3 在不同的细胞核中Cazymes家族基因表达
通过分析Cazymes家族差异基因,在比较组CK3 vs. CK1、WP3 vs. WP1差异表达的基因分别为116和49个(表1),且上调表达基因的数量占优。
表1 在两组比较组中Cazymes家族上调下调情况
Table 1
| CK3 vs. CK1 | WP3 vs. WP1 | |||
|---|---|---|---|---|
| Up | Down | Up | Down | |
| Auxiliary activities (AAs) | 5 | 3 | 17 | 0 |
| Carbohydrate esterases (CEs) | 5 | 3 | 12 | 0 |
| Glycoside hydrolases (GHs) | 15 | 15 | 57 | 7 |
| Glycosyl transferases (GTs) | 1 | 1 | 12 | 7 |
| Polysaccharide lyases (PLs) | 1 | 0 | 4 | 0 |
| Total | 27 | 22 | 102 | 14 |
进一步分析纤维素酶和木质素酶基因的表达,发现在WP3 vs. WP1和CK3 vs. CK1组中,只有多铜氧化酶、锰过氧化物酶、β-葡萄糖苷酶和内切β-1,4-葡聚糖酶表达有差异(表2),1,4-β-纤维素酶、木质素过氧化物酶和通用过氧物酶表达无差异。异核体WP1和WP3的多铜氧化酶表达有6个基因表达有差异,在WP1中Le3001008和Le2000049表达最高,远远高于WP3,而WP3中Le3000250表达最高达到了935.57,是WP1的5倍。锰过氧化物酶都表现为WP1高于WP3;β-葡聚糖苷酶有5个基因表达有差异,WP1有2个高表达,3个低表达,特别是Le2000265,是WP3的30倍以上;内切β-1,4-葡聚糖酶有3个表达差异,WP3有2个高表达,WP1有1个高表达,且Le5000298在WP3中表达最高。进一步比较CK1 vs. CK3,多铜氧化酶有3个基因表达有差异,且全部为CK1全部上调,表达最高的为Le3000250;锰过氧化物酶表达差异基因有3个,全部表现为CK1高于CK3;β-葡聚糖苷酶有2个基因表达差异,且都为CK1上调;CK1的3个内切β-1,4-葡聚糖酶基因表达高于CK3 (表2)。
表2 不同异核体中木质纤维素降解酶基因表达情况
Table 2
| Genes | Gene_id | WP3 | WP1 | CK1 | CK3 | WP3 vs. WP1 | CK3 vs. CK1 |
|---|---|---|---|---|---|---|---|
| 多铜氧化酶 Multicopper oxidase | Le3001008 | 0.22±0.15 | 311.05±253.88 | 91.19±36.19 | 0.12±0.14 | Yes|Up | Yes|Up |
| Le1001852 | 0.38±0.07 | 39.58±25.90 | 0.14±0.17 | 0.03±0.05 | Yes|Up | ||
| Le5001036 | 13.09±1.31 | 1.32±0.69 | 3.20±0.54 | 2.06±0.82 | Yes|Down | ||
| Le2000806 | 19.03±1.89 | 8.36±3.67 | 11.86±2.21 | 3.38±0.62 | Yes|Up | ||
| Le2000049 | 19.92±1.13 | 655.97±417.41 | 55.54±35.11 | 39.13±1.48 | Yes|Up | ||
| Le6000624 | 51.09±8.52 | 117.65±30.49 | 59.50±7.93 | 20.27±5.39 | Yes|Up | Yes|Up | |
| Le3000250 | 935.57±34.93 | 188.46±47.04 | 123.00±57.96 | 51.82±12.71 | Yes|Down | ||
| 锰过氧化物酶 Manganese peroxidase | Le3000125 | 14.38±0.73 | 52.82±47.44 | 11.48±0.88 | 4.08±0.16 | Yes|Up | Yes|Up |
| Le2000852 | 3.13±0.69 | 18.30±5.86 | 7.77±5.27 | 0.76±0.19 | Yes|Up | Yes|Up | |
| Le2000871 | 9.14±1.34 | 320.34±340.37 | 30.02±20.24 | 2.87±0.32 | Yes|Up | Yes|Up | |
| β-葡萄糖苷酶 β-glucosidase | Le2000265 | 33.71±6.41 | 1011.49±124.79 | 281.73±32.01 | 72.59±9.01 | Yes|Up | Yes|Up |
| Le2001102 | 10.07±2.01 | 39.79±1.55 | 45.15±4.80 | 6.11±0.44 | Yes|Up | Yes|Up | |
| Le10000333 | 11.10±0.89 | 0.16±0.20 | 2.56±0.45 | 2.45±0.48 | Yes|Down | ||
| Le7000070 | 11.67±0.93 | 0.51±0.22 | 2.29±0.51 | 2.27±0.12 | Yes|Down | ||
| Le5000652 | 25.59±0.46 | 2.26±1.45 | 10.53±1.79 | 8.03±1.18 | Yes|Down | ||
| 内切β-1,4-葡聚 糖酶 Endo-beta- 1,4-glucanase | Le5000298 | 113.29±7.81 | 33.51±5.42 | 26.13±2.64 | 44.80±6.92 | Yes|Down | |
| Le4000095 | 3.47±0.69 | 4.82±4.22 | 25.71±5.54 | 4.99±1.42 | Yes|Up | ||
| Le8000310 | 37.00±2.48 | 2.34±1.50 | 9.48±2.53 | 5.88±1.59 | Yes|Down | ||
| Le6000389 | 5.34±1.86 | 11.79±6.16 | 37.61±9.77 | 6.95±1.43 | Yes|Up | Yes|Up | |
| Le4000845 | 5.57±0.84 | 1.94±0.48 | 16.77±2.93 | 1.32±0.37 | Yes|Up |
3 讨论
研究发现香菇单菌丝的生长速度与交配型具有相关性,不同交配型的生长速度以及菌落形态有较大的差异(程水明 2005;林范学等 2013;程爽爽等 2019),这些差异的产生可能与其遗传背景相关,通过分析发现香菇双核菌株的异核体的基因组存在着遗传背景差异大的问题,包括大段的染色体重排、同源染色体间的长度差异较大、Cazymes数量和基因功能以及代谢通路上都存在差异。通过本研究中异核体转录组的分析发现,2个异核菌株的表达谱同样存在差异,从细胞代谢到细胞结构等途径都存在差异,其中碳水化合物和氨基酸代谢等都得到了富集,特别是Y0040-1比Y0040-3菌株的Cazymes家族,包括纤维素酶和木质素酶上调基因占优,这说明Y0040-1可能潜在的木质纤维素降解能力较强。在双孢蘑菇中发现其中一个核的代谢相关基因和Cazymes家族的基因表达占优势,是导致异核体生长不同的原因(Thies et al. 2018),基于此可能也是Y0040-1比Y0040-3生长速度快的原因。
参考文献
Research progress on the mating-typing locus structures of basidiomycete mushrooms
Study on the phenomenon and genetic basis of segregation distribution of mating-type factors in Lentinus edodes
Analysis of mycelial growth characteristics of Lentinus edodes monokaryons strains
The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes
DOI:10.1126/science.1221748
PMID:22745431
[本文引用: 1]

Wood is a major pool of organic carbon that is highly resistant to decay, owing largely to the presence of lignin. The only organisms capable of substantial lignin decay are white rot fungi in the Agaricomycetes, which also contains non-lignin-degrading brown rot and ectomycorrhizal species. Comparative analyses of 31 fungal genomes (12 generated for this study) suggest that lignin-degrading peroxidases expanded in the lineage leading to the ancestor of the Agaricomycetes, which is reconstructed as a white rot species, and then contracted in parallel lineages leading to brown rot and mycorrhizal species. Molecular clock analyses suggest that the origin of lignin degradation might have coincided with the sharp decrease in the rate of organic carbon burial around the end of the Carboniferous period.
Optimization of culture conditions for Lentinus sp.protoplast fusant
The gene expression profiles of promoting mycelial growth of monokaryotic strains of Lentinula edodes cultured on PDA with additional sawdust
A study on relative dominance of the nuclei during protoplast monokaryotization of Lentinula edodes
Identification of secondary recombinants and linked molecular markers for mating-type factors in Lentinula edodes
Protoplast monokaryogenesis and cross of the homokaryotic strains of Wolfiporia hoelen
The relationships between mating types and mycelial growth rates of monokaryons and dikaryons in Lentinula edodes
Genetic structure and polymorphism of mating-type loci in different Hypsizygus marmoreus strains
Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche
Optimum of stock culture medium formula of Auricularia fuscosuccinea by orthogonal test
Variance analysis of different mating type strains in mononuclear protoplast of Hypsizygus marmoreus
Comparative transcriptome analysis identified candidate genes involved in mycelium browning in Lentinula edodes
Differential expression analysis for sequence count data
Comparative transcriptome analysis of dikaryotic mycelia and mature fruiting bodies in the edible mushroom Lentinula edodes
DOI:10.1038/s41598-018-27318-z
[本文引用: 1]

Lentinula edodes is a popular cultivated edible mushroom with high nutritional and medicinal value. To understand the regulation of gene expression in the dikaryotic mycelium and mature fruiting body in the commercially important Korean L. edodes strain, we first performed comparative transcriptomic analysis, using Illumina HiSeq platform. De novo assembly of these sequences revealed 11,675 representative transcripts in two different stages of L. edodes. A total of 9,092 unigenes were annotated and subjected to Gene Ontology, EuKaryotic Orthologous Groups, and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Gene expression analysis revealed that 2,080 genes were differentially expressed, with 1,503 and 577 upregulated in the mycelium and a mature fruiting body, respectively. Analysis of 18 KEGG categories indicated that fruiting body-specific transcripts were significantly enriched in ‘replication and repair’ and ‘transcription’ pathways, which are important for premeiotic replication, karyogamy, and meiosis during maturation. We also searched for fruiting body-specific proteins such as aspartic protease, gamma-glutamyl transpeptidase, and cyclohexanone monooxygenase, which are involved in fruiting body maturation and isolation of functional substances. These transcriptomes will be useful in elucidating the molecular mechanisms of mature fruiting body development and beneficial properties, and contribute to the characterization of novel genes in L. edodes.
Innovation of Lentinus edodes germplasm by protoplast monokaryon hybridization technology
Breeding of protoplast mononuclear hybrid strain “LNX” series of Lentinus edodes
Nucleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regulatory programs
Selection and breeding for new strains in Lentinula edodes
Comparative genomic analysis of lignocellulolytic enzymes in different Lentinula edodes strains
Study on the differences of monokaryotic mycelium and dikaryotic mycelium of Ganoderma lucidum
Transcriptome analysis identified candidate genes involved in fruit body development under blue light in Lentinula edodes
DOI:10.3390/app11156997
URL
[本文引用: 1]

Lentinula edodes is an edible mushroom that is rich in polysaccharides, glucan, and lentinan. It is famous for its earthy, sweet, umami flavor, and is used in various foods all over the world. Although Lentinula edodes does not carry out photosynthesis with light, its fruit body development is regulated by light. In this study, we analyzed the morphological changes of L. edodes strain Sanjo701ho and identified the global gene expression patterns using EdgeR of fruit body development under blue light. The phenotype analysis under different light sources revealed that the pileus diameter grew, while the stipe length was suppressed under blue light. To understand the changes in the transcriptome under different light sources in L. edodes, gene set enrichment analysis (GSEA), KOG functional categories, and KEGG pathways were used and compared to the no-light condition. Lignocellulose, CAZyme, and transcription factor classified DEGs (differentially expressed genes) were identified to better understand the significant DEGs affected by light sources in the synthesis, metabolism, and recognition of complex carbohydrates. Six glycoside hydrolases (GHs), four auxiliary activities (AAs), three carbohydrate esterases (CEs), and glycosyltransferases (GTs) were identified as upregulated in the CAZyme DEGs. Furthermore, four β-glucosidase, one glucose oxidase, and one multicopper oxidase-related gene for lignocellulolytic genes were upregulated in the blue light condition, and AT_hook transcription factor, CBFD_NFYB_HMF transcription factor, HMG_box transcription factor, and fungal specific transcription factor were upregulated in the blue light condition. This study helps us understand fruit body development in mushroom-breeding programs.
Isolation and comparison of different types of protoplast monokaryons from Flammulina velutipes
茯苓原生质体单核化及同核体杂交
香菇原生质体单核杂交菌株“LNX”系列的选育初报
中国菌物学会2016年学术年会, 福州. 128