瓶口衣科内的鳞核衣类地衣(catapyrenioid lichens)是指地衣体鳞片状的类群,包括广义鳞核衣Catapyrenium s.l.和石果衣属Endocarpon Hedw. (Prieto et al. 2010a, 2012),前者的共有表型特征是子囊孢子单胞型,子实层无共生藻(Breuss 2010),与子囊孢子砖壁型、子实层存在共生藻的石果衣属明显区分。广义鳞核衣包括8个属,其中的盾鳞衣属Placidium A. Massal.指地衣体鳞片型、髓层与下皮层发育良好、子囊棍棒形或者圆柱形、分生孢子器Dermatocarpon型、裂片表面生(laminal)或者边缘生(marginal)的类群。盾鳞衣属现包括31种(Breuss 2010;Prieto et al. 2012;Usman et al. 2021;Zhang et al. 2022),中国已报道5种(Wei 2020;Zhang et al. 2022)。石果衣属指地衣体鳞片型、有复杂的假根、子囊壳单个沉浸式、子实层有藻、侧丝交联在一起、子囊孢子砖壁型的类群(Breuss 2002),已知55-65种,中国已报道14种(Aptroot 2003;Harada et al. 2016;Wei 2020)。
盾鳞衣属和石果衣属地衣的生长基物包括岩石、沙土、树皮,分布范围广泛。我国幅员辽阔、生态环境多样,荒漠地区的瓶口衣科物种数目被严重低估。为澄清中国石果衣属和盾鳞衣属物种多样性,本研究基于形态学特征和分子系统学分析,对保存在中国科学院微生物研究所菌物标本馆地衣标本室(HMAS-L)的近几年采集的两属标本进行了整理和鉴定,发现1个新种和3个新记录种。
1 材料与方法
1.1 研究标本采集和形态特征观察
研究标本于2019-2022年采集自甘肃、青海、新疆、河南4个省和自治区,大部分属于干旱、半干旱的荒漠地区,现保存在中国科学院菌物标本馆地衣标本室(HMAS-L)。在LEICA M125体式显微镜下观察地衣体的外表特征,并用显微镜自带的Leica DFC450相机拍照。徒手结合Leica CM1950冷冻切片机切片,选取10-15 µm厚的鳞片或子实体薄片,在配备有Zeiss AxioCam MRC5相机的Zeiss Axio Imager A2-M2显微镜观察解剖特征并拍照。
1.2 地衣化学物质检测
因文献报道石果衣属和盾鳞衣属的物种不产生次级代谢产物,因此本研究对标本只根据Brodo (2003)描述的薄层层析法进行检验,所用的溶剂为C系统(甲苯:乙酸=20:3,体积比)。
1.3 DNA提取、PCR扩增和测序
使用体式显微镜MOTIC SMZ-168操作选取形态一致的地衣体鳞片或子囊盘,去除附着的基物后使用无菌水与75%乙醇涡旋清洗。标本总的DNA提取使用改良的CTAB法(Rogers & Bendich 1989),ITS区使用引物ITS4和ITS5进行PCR扩增(White et al. 1990),nuLSU扩增使用引物PRI1与PRI2 (Prieto et al. 2010b),RPB1扩增使用引物RPB1-1F和RPB1-6R1asc (Gueidan et al. 2007)。扩增的体系和程序参照Gueidan et al. (2007)、Prieto et al. (2010b)和Zhang et al. (2022)的方法进行,以上扩增产物经1%琼脂糖凝胶检验后产物送北京天一辉远生物科技有限公司纯化和测序。
1.4 序列比对和系统发育分析
测序获得的单端序列使用SeqMan (Swindell & Plasterer 1997)进行校正和拼接,在GenBank中下载构建系统发育树所需要的序列,使用MAFFT v. 7 (Katoh et al. 2009)进行多重序列比对,使用Gblocks V0.19b (Castresana 2000;Talavera & Castresana 2007)来去除比对产生的模糊序列。单基因片段经PAUP* v. 4.0 (Swofford 2002)同质性检验符合后(P>0.05)联合获得多基因矩阵。最大似然法(maximum likelihood, ML)和贝叶斯法(Bayes)分析在Cipres Science Gateway (
2 结果与分析
2.1 分子系统学分析结果
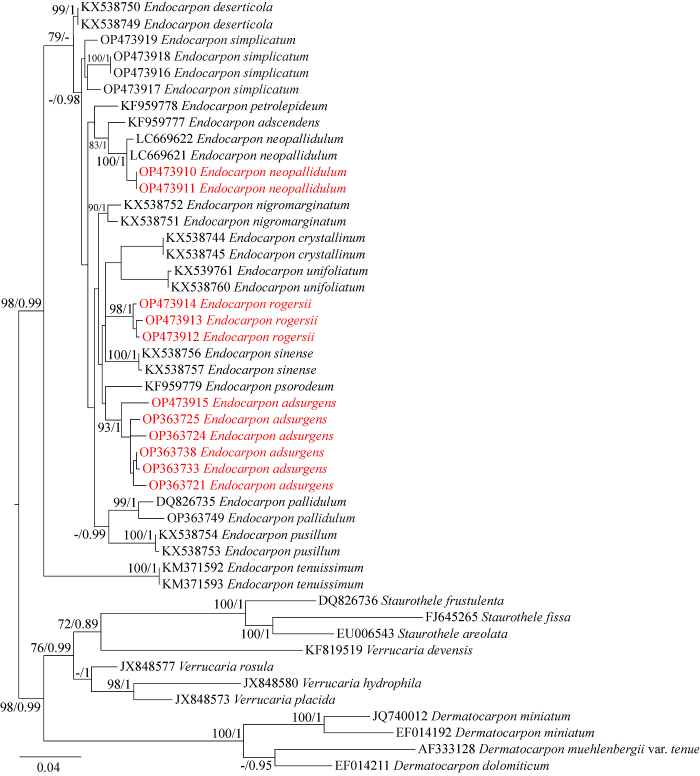
本研究石果衣属新产生16条ITS序列,已提交GenBank,与下载的32条参考序列(附表1,国家微生物科学数据中心NMDCX0000174)比对、去除模糊序列后形成包括25个物种的491个位点矩阵。比对ML树与BI树,有相同的拓扑结构,因此文中仅展示ML树,将BI树的后验概率标记在节点的分支处的自展值之后,BI树见附图1 (国家微生物科学数据中心NMDCX0000174)。本文描述的5个物种在进化树上各自形成高支持率的分支(图1)。
图1
图1
基于ITS序列的最大似然树
每个分支的数值表示自展值(BS)和贝叶斯后验概率(PP),仅显示自展值大于75或后验率大于0.95的数值. 该系统发育树中瓶口衣科的矮疣衣属、瓶口衣属和皮果衣属为外群. 红色标记的序列由本研究新产生. 核苷酸替代率为0.04
Fig. 1
The RAxML tree based on the concatenated ITS data set representing both ML and BI trees.
The number in each node represents bootstrap support (BS) and posterior probability (PP) values. BS values≥75 and PP values≥0.95 were plotted on the branches. The phylogenetic tree was rooted to Staurothele spp., Verrucaria spp., and Dermatocarpon spp. of Verrucariaceae. The sequences of taxa in red color were newly generated for this study. Scale in 0.04 substitution per site.
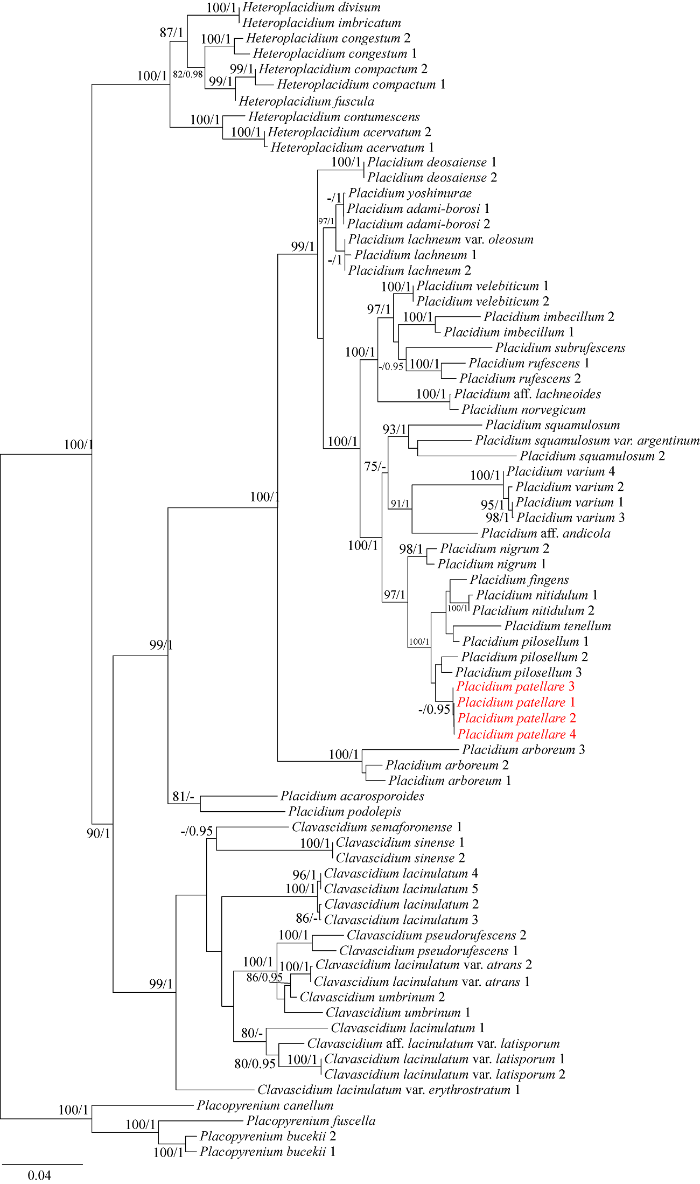
本研究中盾鳞衣属共新产生16条序列(4 ITS, 2 nuLSU, 10 RPB1),与参考序列共118条DNA序列(附表2,国家微生物科学数据中心NMDCX0000174)用于系统分析。多位点比对、去除模糊序列后形成包括75个物种的2 262个位点矩阵(483 ITS, 780 nuLSU和999 RPB1)。比对ML树与BI树,有相同的拓扑结构,文中仅展示ML树,将BI树的后验概率标记在节点的分支处的自展值之后(图2),BI树与单基因树见附图2-5 (国家微生物科学数据中心NMDCX0000174)。
图2
图2
基于ITS+nuLSU+RPB1序列的最大似然树
每个分支的数值表示自展值(BS)和贝叶斯后验概率(PP),仅显示自展值大于75或后验概率大于0.95的数值. 该系统发育树以瓶口衣科的叶核衣属为外群. 红色标记的为本研究报道的新种. 核苷酸替代率0.04
Fig. 2
The RAxML tree based on the concatenated ITS+nuLSU+RPB1 data set.
The numbers in each node represent bootstrap support (BS) and posterior probability (PP) values. BS values≥75 and PP values≥0.95 were plotted on the branches of the tree. The phylogenetic tree was rooted to Placopyrenium spp. of Verrucariaceae. The samples corresponding to the new species are in red color. Scale in 0.04 substitution per site.
本研究中的新物种已在Fungal Names网站(
2.2 物种描述
2.2.1 卷边石果衣 图3
Endocarpon adsurgens Vain., Acta Soc. Fauna Flora Fenn. 49 (no. 2): 73 (1921). Fig. 3
图3
图3
卷边石果衣
A:地衣体外表(HMAS-L 154868). B:地衣体外表(HMAS-L 154850). C:假根 (HMAS-L 154868). D:鳞片纵切解剖图(HMAS-L 154868). E:假根纵切解剖图(HMAS-L 154868). F:子囊壳纵切结构图(HMAS-L 154868). G-K:子囊与子囊孢子(HMAS-L 154868). 标尺:A=2 mm;B=1 mm;C=2 mm;D=100 μm;E=20 μm;F=100 μm;G-K=10 μm
Fig. 3
Endocarpon adsurgens.
A, B: The habit of squamulose thallus. C: Rhizines. D: Transversal section of thallus. E: Transversal section of rhizines. F: Transversal section of perithecium. G-K: Asci and ascospores. Bars: A=2 mm; B=1 mm; C=2 mm; D=100 μm; E=20 μm; F=100 μm; G-K=10 μm.
形态特征:地衣体鳞片状,黄棕色至深棕色,表面具蜡质光泽,有时具有较多的黑色小点,为成熟的子囊壳;裂片直径1-4 (-5) mm,紧贴基物生长,边缘浅裂,多上卷,幼时平坦;下表面深黑色,与边缘同色,具有发达的黑色假根。
解剖特征:裂片有时具有淡灰色、透明的外层,厚20 µm;外层与上皮层之间称为最上层的厚度8-10 µm,黄棕色;上皮层厚45-60 µm,灰色;藻层连续,厚55-70 µm,由直径5 µm的近球形绿藻细胞组成;下皮层深灰棕色,厚45-50 µm;髓层深棕色,厚245-355 µm;假根菌丝束宽100-200 µm,由棕色的假根菌丝组成,单根菌丝直径4-5 µm。分生孢子器未见。
子实体:子囊壳完全沉浸入地衣体,宽梨形,300×350 µm,成熟时外壁黑色,厚30-50 µm,顶端开口处增厚,达60 µm,无包顶组织(involucrellum);侧丝不分枝,长25-30 µm;子实层厚65-96(-115) µm,浅灰棕色;子囊(75-90)×(18-20) µm,内含2个孢子;子囊孢子砖壁型,长椭圆形到亚棍棒形,淡灰棕色,透明,(35-45)×(13-15) (-20) µm。
化学成分:TLC未检测到化学物质。
生长基物:沙土。
分布:美国西北部的蒙大拿州、芬兰、俄罗斯、奥地利(Breuss & McCune 1994)。该种为首次在中国报道,在我国分布范围较广,青藏高原、西北地区沙漠边缘均有分布。
研究标本:甘肃:白银市,景泰县翠柳沟山谷内,37°25′06″N,104°34′56″E,1 591 m alt.,沙土,2020.10.21,钱旭等20201499 (HMAS-L 154868),20201473 (HMAS-L 154850);玉门市,老玉门镇废弃油田内野山上,39°48′36″N,97°17′02″E,2 354 m alt.,沙土,2020.10.18,钱旭等20201454 (HMAS-L 154838)。青海:海南藏族自治州,兴海县207国道旁的野山上,35°42′31″N,100°14′51″E,3 004 m alt.,沙土,2020.10.16,钱旭等20201375 (HMAS-L 154768);贵南县尕玛羊曲黄河大桥旁的野山上,35°40′54″N,100°15′08″E,1 606 m alt.,沙土,2020.10.16,钱旭等20201382 (HMAS-L 154803);共和县恰卜恰镇祁连山金河水泥有限公司对面的土坡上,36°15′45″N,100°34′28″E,2 831 m alt.,沙土,2020.10.16,钱旭等20201391 (HMAS-L 154806)。
讨论:该种地衣体外形与Endocarpon pusillum Hedw.相似,下表面都生长有黑色假根,区别在于E. pusillum的鳞片边缘更圆且紧贴基物生长,假根相比该种更脆弱。Endocarpon adscendens Malme与E. adsurgens相比,下表面呈中间黑色过渡到边缘灰白色,无假根。在解剖形态上,E. adscendens和E. pusillum的子囊孢子均为深棕色,而E. adsurgens孢子偏灰色(Breuss & McCune 1994)。
2.2.2 新白色石果衣 图4
Endocarpon neopallidulum H. Harada, Nova Hedwigia 56(3-4): 340 (1993). Fig. 4
图4
图4
新白色石果衣(HMAS-L 154966)
A,B:地衣体外表. C:鳞片纵切解剖图. D,E:子囊壳纵切解剖图. F:鳞片纵切结构图. G-I:子囊与子囊孢子. 标尺:A=1 mm;B=0.5 mm;C=200 μm;D,E=50 μm;F-I=20 μm.
Fig. 4
Endocarpon neopallidulum (HMAS-L 154966).
A, B: Habit of thallus. C: Transversal section of thallus. D, E: Transversal section of perithecium. F: Transversal section of thallus. G-I: Asci and ascospores. Bars: A=1 mm; B=0.5 mm; C=200 μm; D, E=50 μm; F-I=20 μm.
形态特征:地衣体鳞片型,紧贴基物生长,偶见重叠覆盖;上表面橄榄绿色,黯淡无光,有深棕色的成熟子囊壳,略微突起,最高0.1 mm;裂片边缘圆润或浅裂,宽0.2-0.8 mm;下表面灰白色,无假根菌丝束。
解剖特征:裂片很薄,厚100-150 µm;外层未见,最上层浅棕色,厚8-10 µm;上皮层很薄,不易见,厚10-15 µm;藻层连续,厚70-75 µm,几乎弥漫了整个裂片纵切面,由直径6-8 µm的近球形绿藻细胞组成,绿藻细胞每2-4个聚集在一起;髓层混合型,由球形和延长的丝状细胞组成,淡灰色,厚约40 µm;下皮层深棕色,厚约25-40 µm。分生孢子器未见。
子实体:子囊壳宽梨形,直径约140-150 µm,成熟时外壁黑色,厚28-32 µm,无包顶组织;侧丝不分枝,长18-22 µm;子实层厚25-30 µm,浅灰棕色;子囊60-70×15-20 µm,内含两个孢子;子囊孢子砖壁型,长椭圆形至亚棍棒形,淡黄色透明,33-40×13-18(-20) µm。
化学成分:TLC未检测到化学物质。
生长基物:岩石。
分布:该种仅在日本报道过(Harada 1993)。本研究中的2份标本采集自位于亚热带的大别山区域。
研究标本:河南:信阳市,光山县大苏山国家森立公园内靠近净居寺茶场,31°52′03″N,114°48′22″E,106 m alt.,岩石,2022.7.21,魏鑫丽等20220599 (HMAS-L 154965),20220600 (HMAS-L 154966)。
2.2.3 罗杰氏石果衣 图5
Endocarpon rogersii P.M. McCarthy, Lichenologist 23(1): 43 (1991). Fig. 5
图5
图5
罗杰氏石果衣(HMAS-L 143991)
A,B:地衣体外表与裂片表面成熟的子囊壳开口. C:子囊壳纵切解剖图. D:子囊壳开口处的侧丝. E:分生孢子器纵切解剖图. F:分生孢子. G-I:子囊孢子. 标尺:A,B=1 mm;C=100 μm;D=20 μm;E=100 μm;F=20 μm;G=10 μm;H-I=20 μm
Fig. 5
Endocarpon rogersii (HMAS-L 143991).
A, B: Habit of thallus. C: Transversal section of perithecium. D: Paraphyses. E: Transversal section of pycnidium. F: Conidia. G-I: Ascospores. Bars: A, B=1 mm; C=100 μm; D=20 μm; E=100 μm; F=20 μm; G=10 μm; H-I=20 μm.
形态特征:地衣体鳞片型,紧贴基物生长,未见重叠覆盖;上表面灰棕色,具有灰白色粉霜,黯淡无光,有深棕色的成熟子囊壳,宽0.2-0.25 mm;鳞片圆润或边缘深裂,宽1-4 mm;下表面灰棕色,假根与下表面同色。
解剖特征:裂片厚200-250 µm;外层未见,最上层浅棕色,厚20 µm;上皮层很薄,灰色,厚18-24 µm;藻层连续,厚80-110(-130) µm,由直径6-8 µm的近球形绿藻细胞组成。髓层假薄壁型,灰棕色,厚约75-95(-120) µm;下皮层缺失,髓层直接连接直径4-5 µm的淡黄棕色假根菌丝。分生孢子器形状不规则,宽(-100)150-350(-450) µm,黄棕色,沉浸入裂片中;分生孢子无色,长杆状,1-1.5×(-3.5)4.5-6.5 (-7.5) µm。
子实体:子囊壳近圆形,直径约250-300 µm,成熟时外壁深棕色,开口处变厚,厚45-60 (-80) µm,无包顶组织;侧丝不分枝,长30-40 µm;子实层厚45-50 µm,浅黄棕色。子囊70-90× 20-25 µm,内含两个孢子;子囊孢子砖壁型,长椭圆形至亚棍棒形,从透明黄棕色到不透明深棕色,30-40(-45)×15-20 µm。
化学成分:TLC未检测到化学物质。
生长基物:沙土。
分布:该种分布在年降水不超过400 mm的半干旱地区寡营养沙土上,如南澳大利亚的新南威尔士州(McCarthy 2007)。此次为在中国首次报道,在我国的分布地年降水均低于100 mm。
研究标本:新疆:昌吉回族自治州,阜康市梧桐沟中心管护站内,44°22′59″N,87°52′34″E,438 m alt.,沙土,2021.5.9,钱旭和张婷婷20210664 (HMAS-L 151843)。甘肃:武威市,民勤县治沙站内封育绿洲边缘,38°35′08″N,102°58′26″E,1 381 m alt.,沙土,2019.5.3,魏鑫丽20191385 (HMAS-L 143991);治沙站西沙窝,38°34′29″N,102°58′57″E,1 379 m alt.,沙土,2021.5.2,魏鑫丽20191363 (HMAS-L 143990)。
讨论:该物种的显著特征是地衣体上表面具粉霜,裂片边缘深裂,上皮层较薄,假根非常细。与该种相似的物种为E. pallidum,但后者缺乏假根,且裂片宽度不超过2 mm。另一与该种易混淆的是Endocarpon rosettum A. Singh & Upreti,后者地衣体上表面无粉霜,上皮层细胞完全无色且更大,裂片厚度不超过150 µm,子囊壳宽150-200 µm (McCarthy 2007)。
2.2.4 盘形盾鳞衣 图6
Placidium patellare T.T. Zhang & X.L. Wei, sp. nov. Fig. 6
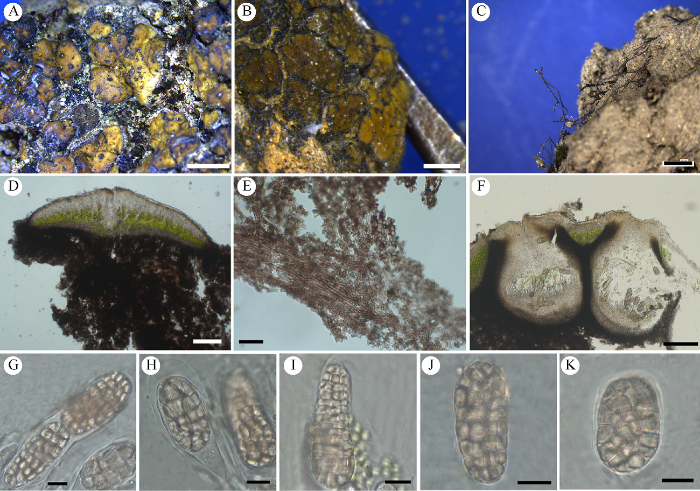
图6
图6
盘形盾鳞衣
A:鳞片状地衣体(HMAS-L 151943). B:鳞片状地衣体(HMAS-L 143914). C:鳞片纵切解剖图(HMAS-L 151943). D:鳞片纵切解剖图(HMAS-L 143914). E:沉浸入鳞片的子囊壳(HMAS-L 143914). F:边缘生的分生孢子器(HMAS-L 143914). G:圆柱状子囊内单列排布的孢子(HMAS-L 151943). H:棒状子囊内双列排布的孢子(HMAS-L 143914). I:分生孢子(HMAS-L 143914). 标尺:A,B=2 mm;C,D=50 µm;E=200 µm;F=100 µm;G,H=20 µm;I=10 µm
Fig. 6
Placidium patellare.
A: Squamulose thallus (HMAS-L 151943). B: Squamulose thallus (HMAS-L 143914). C: Transversal section of thallus (HMAS-L 151943). D: Transversal section of thallus (HMAS-L 143914). E: Immersed perithecia (HMAS-L 143914). F: Marginal pycnidium (HMAS-L 143914). G: Uniseriate ascospores in cylindrical asci (HMAS-L 151943). H: Biseriate ascospores in clavate asci (HMAS-L 143914). I: Conidia (HMAS-L 143914). Bars: A, B=2 mm; C, D=50 µm; E=200 µm; F=100 µm; G, H=20 µm; I=10 µm.
Fungal Names: FN571249
Diagnosis: This new species is different from the similar species Placidium acarosporoides in its terricolous habit, flatter squamules, marginal pycnidia, and smaller ascospores.
Etymology: The epithet ‘patellare’ refers to the patellate squamules.
Type: QINGHAI: Haixi Mongolian and Tibetan Autonomous Prefecture, Wulan County. 36°57′31″N, 98°54′03″E, 3 314 m alt., on the sand, 25 April 2021, X.L. Wei & T.T. Zhang 20210515 (holotype, HMAS-L 151943).
Description: Thallus squamulose, terricolous; lobes 1-4 mm wide, (185)231±37(299) µm thick, roundish to slightly lobed, discrete to contiguous, rarely overlapping, full appressed to the substrate or with raised margins free from the substrate; upper surface medium to dark brown, dull, pruinose; lower surface pale to pale yellow; epinecral layer if presence 32-40 µm thick; upper cortex paraplectenchymatous, (18)49±20(78) µm thick, cells (7.8)9.9±1.7(12.6) µm in diam., uppermost layer bright brown, 15-23 µm thick; algal layer (61)88±13(104) µm thick, composed of rounded cells (11.8)13.4±1.6(17.4) µm in diam., globose to sub-globose; medulla mixed type, (34)46±8(58) µm thick; lower cortex clearly delimited from the medulla, transparent, (29)41±7(53) µm thick, composed of rounded cells (7.4)10.9±1.5(13.7) µm diam. Rhizohyphae 4-5 µm thick, colorless. Pycnidia marginal, Dermatocarpon-type, subglobose, light brown, conidia oblong-ellipsoid to bacilliform, 300 µm in diam., (3.4)3.5±0.28(4)×(1.2)1.9±0.32(2.4) µm.
Perithecia laminal, immersed, subglobular (160 µm in diam.) to pyriform (180×240 µm); perithecia wall grey, 20-35 µm thick; hymenium pale brown, 50-74 µm thick; involucrellum not seen; paraphyses 20-35 µm long; asci cylindrical (82-95×11-15 µm) to (sub) clavate (43-52×14- 19 µm), 8 spored, ascospores uniseriate to (sub) biseriate, hyaline, narrow ellipsoid to subglobose, (8.1)10.4±1.6(13.4)×(6.3)7.3±0.6(8.3) µm.
Chemistry: No substances were detected by TLC.
Habitat and distribution: This species colonized on the surface of sandy soil in the semi-arid and arid region of northwest China, with large range of altitude variation from 1 563 m to above 3 300 m. It is only known in China up to now.
Specimens examined: QINGHAI: Haixi Mongolian and Tibetan Autonomous Prefecture, Wulan County. 36°57′31″N, 98°54′03″E, 3 314 m alt., on the sand, 25 April 2021, X.L. Wei & T.T. Zhang 20210501 (HMAS-L 151959); Delhi City. 37°20′27″N, 97°13′00″E, 2 922 m alt., on the sand, 24 April 2021, X.L. Wei & T.T. Zhang 20210403 (HMAS-L 151961). INNER MONGOLIA: Alxa Right Banner, 39°28′22′′N, 101°04′04′′E, 1 563 m alt., on sand, 5 June 2018, D.L. Liu et al. ALS2018015 (HMAS-L 143914).
Notes: This species can be easily distinguished by its patellate squamules, but to some extent, this character is also seen in Placidium acarosporoides (Zahlbr.) Breuss (Nash et al. 2002), which differs in saxicolous habit, bullate squamules, laminal pycnidia, and larger ascospores. Phylogenetic trees (Fig. 2 and National Microbiology Data Center NMDCX0000174 Figs. S2-S5) indicate that the new species is close to Placidium fingens (Breuss) Breuss, Placidium nitidulum T.T. Zhang & X.L. Wei, Pl. tenellum, and Placidium pilosellum (Breuss) Breuss. The first three species have laminal pycnidia, which can be easily distinguished from Pl. patellare. Although Pl. pilosellum also has marginal pycnidia like Pl. patellare, it has hairy-margined squamules (Prieto et al. 2010a) and slender ascospores comparing to Pl. patellare (12-17× 5.5-7.5 µm) (Nash et al. 2002). Although there exist much differences in thallus habit, epinecral layer, and asci among the specimens of Pl. patellare collected from different-altitude sites, the phylogenetic tree well supports its monophyly (Fig. 2 and National Microbiology Data Center NMDCX0000174 Figs. S2-S5). Therefore, combined with the above-mentioned morphological characteristics, we identified Pl. patellare as a new species.
特征提要:该新种与Placidium acarosporoides相似,区别在于该新种土生、裂片较平坦、分生孢子器靠裂片边缘生长和子囊孢子更小。
词源:新种拉丁名种加词‘patellare’指地衣鳞片呈盘形。
主模式:青海:海西蒙古族藏族自治州,德令哈市,乌兰县,S2013 (德兰高速)察汗诺附近的石山。36°57′31″N,98°54′03″E,3 314 m alt.,沙土,2021.4.25,魏鑫丽和张婷婷20210515 (HMAS-L 151943)。
形态特征:地衣体鳞片状,裂片宽1-4 mm,厚(185)231±37(299) µm,中间部分下凹,边缘圆滑或者浅裂,分散或者连续分布,很少见重叠,紧贴基物生长裂片边缘稍翘起;上表面棕色至深棕色,黯淡,有的标本具粉霜;下表面灰白色至淡黄色。解剖特征:外皮层存在但不稳定,厚32-40 µm;上皮层由假薄壁细胞组成,厚(18)49±20(78) µm,细胞直径(7.8)9.9±1.7 (12.6) µm;最上层淡棕色,厚15-23 µm;藻层厚(61)88±13(104) µm,由直径为(11.8)13.4±1.6(17.4) µm的圆形细胞组成;髓层混合型,厚(34)46± 8(58) µm;下皮层与髓层区分明显,透明,厚(29)41±7(53) µm,细胞直径(7.4)10.9±1.5(13.7) µm。假根菌丝宽4-5 µm,无色。分生孢子器靠裂片边缘生长,Dermatocarpon型,近球形,宽达300 µm,浅棕色;分生孢子窄椭圆形到短杆形,(3.4)3.5±0.28(4)×(1.2)1.9±0.32 (2.4) µm。
子实体:子囊壳靠裂片中间生长,沉浸式,较小,近球形(直径达160 µm)或梨形(180×240 µm);子囊壳壁灰色,厚20-35 µm;子实层灰棕色,厚50-74 µm;包顶组织未见;侧丝20-35 µm;子囊圆柱形(82-95×11-15 µm)至(亚)棍棒形(43-52×14-19 µm),子囊8孢;子囊孢子单列到(亚)双列排布,单胞型,窄椭圆形到近球形,无色透明,中心有油滴,(8.1) 10.4±1.6(13.4)×(6.3)7.3±0.6(8.3) µm。
化学成分:TLC未检测到任何化学物质。
生长基物:沙土。
分布:该种分布生境的海拔变化差异大,有3份标本分布在高海拔地区,生于未见明显植被的碎石间的泥土,1份标本分布在腾格里沙漠边缘的山谷中。截至目前该种仅见于中国。
其他研究标本(3份):青海:海西蒙古族藏族自治州,德令哈市,乌兰县,S2013 (德兰高速)察汗诺附近的石山。36°57′31″N,98°54′03″E,3 314 m alt.,沙土,2021.4.25,魏鑫丽和张婷婷20210501 (HMAS-L 151959);西郊克鲁克服务区边的戈壁滩,37°20′27″N 97°13′00″E,2 922 m alt.,沙土,2021.4.21,魏鑫丽和张婷婷20210403 (HMAS-L 151961)。内蒙古蒙古族自治区,阿拉善右旗,西北山中,39°28′22″N,101°04′04″E,1 563 m alt.,沙土,2018.6.5,刘大乐等ALS2018015 (HMAS-L 143914)。
讨论:该新种的标志性特征是裂片圆滑,中心略凹,外形似圆盘。其他具此特征的物种还有Pl. acarosporoides (Nash et al. 2002)。在系统发育树上(Fig. 2和国家微生物科学数据中心NMDCX0000174,Figs. S2-S5),该新种的亲缘关系与Pl. fingens (Breuss) Breuss、Pl. nitidulum T.T. Zhang & X.L. Wei、Pl. tenellum和Pl. pilosellum相近。前3个物种分生孢子器均为靠近裂片中心生长,易与新种区分。Pl. pilosellum的分生孢子器虽然和该新种相似,也是边缘型,但其裂片的边缘呈毛茸状(Prieto et al. 2010a),且子囊孢子更细长(12-17×5.5-7.5 µm) (Nash et al. 2002)。该新种具有一定的遗传多样性,表现在分布的生境、外皮层厚度和子囊形状均有差异,但多基因联合树和单基因树均支持这些差异属于种内范畴(Fig. 2和国家微生物科学数据中心NMDCX0000174 Figs. S2-S5),且Pl. patellare形成的独立分支具高支持(BS和PP值),因此,综合形态和系统发育分析结果,我们认为该新种成立。
致谢
感谢甘肃省治沙研究所刘虎俊研究员在甘肃民勤治沙站协助考察和采集地衣,本课题组博士生钱旭协助采集标本,中国医学科学院医药生物技术研究所张涛副研究员提供文献,中国科学院菌物标本馆地衣标本室(HMAS-L)杨秋霞助理工程师在研究中提供帮助。
参考文献
Pyrenocarpous lichens and related non-lichenized ascomycets from Taiwan
An updated world-wide key to the Catapyrenioid lichens (Verrucariaceae)
DOI:10.13158/heia.23.2.2010.205 URL [本文引用: 2]
Additions to the Pyrenolichen flora of north America
DOI:10.2307/3243901 URL [本文引用: 2]
Microchemical methods for the identification of lichens
DOI:10.1639/0007-2745(2003)106[0345:R]2.0.CO;2 URL [本文引用: 1]
Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis
DOI:10.1093/oxfordjournals.molbev.a026334
PMID:10742046
[本文引用: 1]

The use of some multiple-sequence alignments in phylogenetic analysis, particularly those that are not very well conserved, requires the elimination of poorly aligned positions and divergent regions, since they may not be homologous or may have been saturated by multiple substitutions. A computerized method that eliminates such positions and at the same time tries to minimize the loss of informative sites is presented here. The method is based on the selection of blocks of positions that fulfill a simple set of requirements with respect to the number of contiguous conserved positions, lack of gaps, and high conservation of flanking positions, making the final alignment more suitable for phylogenetic analysis. To illustrate the efficiency of this method, alignments of 10 mitochondrial proteins from several completely sequenced mitochondrial genomes belonging to diverse eukaryotes were used as examples. The percentages of removed positions were higher in the most divergent alignments. After removing divergent segments, the amino acid composition of the different sequences was more uniform, and pairwise distances became much smaller. Phylogenetic trees show that topologies can be different after removing conserved blocks, particularly when there are several poorly resolved nodes. Strong support was found for the grouping of animals and fungi but not for the position of more basal eukaryotes. The use of a computerized method such as the one presented here reduces to a certain extent the necessity of manually editing multiple alignments, makes the automation of phylogenetic analysis of large data sets feasible, and facilitates the reproduction of the final alignment by other researchers.
jModelTest 2: more models, new heuristics and parallel computing
DOI:10.1038/nmeth.2109 PMID:22847109 [本文引用: 1]
Using a multigene phylogenetic analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota)
Verrucariaceae are a family of mostly crustose lichenized ascomycetes colonizing various habitats ranging from marine and fresh water to arid environments. Phylogenetic relationships among members of the Verrucariaceae are mostly unknown and the current morphology-based classification has never been confronted to molecular data. A multilocus phylogeny (nuLSU, nuSSU and RPB1) was reconstructed for 83 taxa representing all main genera of this family to provide a molecular phylogenetic framework necessary to assess the current morphology-based classification. Four main well-supported monophyletic groups were recovered, one of which contains seven robust monophyletic subgroups. Most genera, as traditionally delimited, were not monophyletic. A few taxonomic changes are proposed here to reconcile the morphology-based classification with the molecular phylogeny (Endocarpon diffractellum comb. nov., Heteroplacidium fusculum comb. nov., and Bagliettoa marmorea comb. nov.). Ancestral state reconstructions show that the most recent common ancestor of the Verrucariaceae was most likely crustose with a weakly differentiated upper cortex, simple ascospores, and hymenium free of algae. As shown in this study, the use of symplesiomorphic traits to define Verrucaria, the largest and type genus for the Verrucariaceae, as well as the non monophyly of the genera Polyblastia, Staurothele and Thelidium, explain most of the discrepancies between the current classification based on morphological similarity and a classification using monophyly as a grouping criterion.
A taxonomic study of the lichen genus Endocarpon (Verrucariaceae) in Japan
Lichen flora in the arid valley of Jingsha-jiang R., China (2), genus Endocarpon Hedw. (Verrucariaceae)
MRBAYES: Bayesian inference of phylogenetic trees
DOI:10.1093/bioinformatics/17.8.754
PMID:11524383
[本文引用: 1]

The program MRBAYES performs Bayesian inference of phylogeny using a variant of Markov chain Monte Carlo.MRBAYES, including the source code, documentation, sample data files, and an executable, is available at http://brahms.biology.rochester.edu/software.html.
Multiple alignment of DNA sequences with MAFFT
The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota-approaching one thousand genera
DOI:10.1639/0007-2745-119.4.361 URL [本文引用: 1]
The lichen genus Endocarpon Hedwig in Australia
DOI:10.1017/S0024282991000087
URL
[本文引用: 2]

The status of the lichen genus Endocarpon Hedwig (Verrucariaceae) in Australia is reviewed. Ten taxa are recognized including the following newlydescribed entities: E. aridum McCarthy, E. crassisporum McCarthy & R. Filson, E. macrosporum McCarthy, E. robustum McCarthy, E. rogersii McCarthy and E.simplicatum var. bisporum McCarthy. Endocarpon victoriae Müll. Arg. is a synonym of E. simplicatum (Nyl.) Nyl. Endocarpon pallidum Ach. is reported for the first time from Australia. A problematical corticolous specimen from Victoria is discussed.
The genus Catapyrenium s. lat. (Verrucariaceae) in the Iberian Peninsula and the Balearic Islands
DOI:10.1017/S0024282910000319
URL
[本文引用: 3]

A taxonomic treatment of the genera included inCatapyreniums. lat. in the Iberian Peninsula and the Balearic Islands is provided, based on study ofc. 2000 specimens from both herbaria and fresh material collected by the authors from 2005 to 2009 in numerous localities.
Molecular phylogeny of Heteroplacidium, Placidium, and related catapyrenioid genera (Verrucariaceae, lichen-forming Ascomycota)
DOI:10.3732/ajb.1100239
PMID:22210842
[本文引用: 2]

Verrucariaceae is a fascinating lineage of lichenized fungi for which generic and species delimitation is problematic due to the scarcity of discriminating morphological characters. Members of this family inhabit rocks, but they further colonize soils, barks, mosses, and other lichens. Our aim is to contribute to the DNA-based inference of the Verrucariaceae tree of life and to investigate characters that could be useful for proposing a more natural classification. We focused on catapyrenioid genera, which are often part of biological soil crusts, a cryptogam-dominated ecosystem contributing to soil formation and stabilization in arid environments. Understanding their evolution and taxonomy is essential to assess their roles in these fragile and important ecosystems.A multigene phylogeny of Verrucariaceae including catapyrenioid genera is presented. We further examined the phylogenetic relationships among members of Heteroplacidium and Placidium. The evolution of selected characters was inferred using the latter phylogeny.Anthracocarpon and Involucropyrenium were closely related to Endocarpon. Placidium comprised two monophyletic clades sister to Heteroplacidium. Inferred ancestral states of diagnostic characters revealed that the type of medulla and the pycnidia location were homoplasious within the Placidium clade. In contrast, the presence of rhizines was a synapomorphy for Clavascidium.Our results provide new information on the usefulness of characters for delineating groups in Verrucariaceae. Taxonomic changes are proposed to reflect more natural groupings: Heteroplacidium podolepis is transferred to Placidium, and Clavascidium is recognized as a different genus. Eight new combinations are proposed for Clavascidium.
Phylogenetic study of Catapyrenium s. str. (Verrucariaceae, lichen-forming Ascomycota) and related genus Placidiopsis
DOI:10.3852/09-168 URL [本文引用: 2]
Posterior summarization in Bayesian phylogenetics Using Tracer 1.7
DOI:10.1093/sysbio/syy032
PMID:29718447
[本文引用: 1]

Bayesian inference of phylogeny using Markov chain Monte Carlo (MCMC) plays a central role in understanding evolutionary history from molecular sequence data. Visualizing and analyzing the MCMC-generated samples from the posterior distribution is a key step in any non-trivial Bayesian inference. We present the software package Tracer (version 1.7) for visualizing and analyzing the MCMC trace files generated through Bayesian phylogenetic inference. Tracer provides kernel density estimation, multivariate visualization, demographic trajectory reconstruction, conditional posterior distribution summary, and more. Tracer is open-source and available at http://beast.community/tracer.
MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space
DOI:10.1093/sysbio/sys029
PMID:22357727
[本文引用: 1]

Since its introduction in 2001, MrBayes has grown in popularity as a software package for Bayesian phylogenetic inference using Markov chain Monte Carlo (MCMC) methods. With this note, we announce the release of version 3.2, a major upgrade to the latest official release presented in 2003. The new version provides convergence diagnostics and allows multiple analyses to be run in parallel with convergence progress monitored on the fly. The introduction of new proposals and automatic optimization of tuning parameters has improved convergence for many problems. The new version also sports significantly faster likelihood calculations through streaming single-instruction-multiple-data extensions (SSE) and support of the BEAGLE library, allowing likelihood calculations to be delegated to graphics processing units (GPUs) on compatible hardware. Speedup factors range from around 2 with SSE code to more than 50 with BEAGLE for codon problems. Checkpointing across all models allows long runs to be completed even when an analysis is prematurely terminated. New models include relaxed clocks, dating, model averaging across time-reversible substitution models, and support for hard, negative, and partial (backbone) tree constraints. Inference of species trees from gene trees is supported by full incorporation of the Bayesian estimation of species trees (BEST) algorithms. Marginal model likelihoods for Bayes factor tests can be estimated accurately across the entire model space using the stepping stone method. The new version provides more output options than previously, including samples of ancestral states, site rates, site d(N)/d(S) rations, branch rates, and node dates. A wide range of statistics on tree parameters can also be output for visualization in FigTree and compatible software.
RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies
Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments
DOI:10.1080/10635150701472164
PMID:17654362
[本文引用: 1]

Alignment quality may have as much impact on phylogenetic reconstruction as the phylogenetic methods used. Not only the alignment algorithm, but also the method used to deal with the most problematic alignment regions, may have a critical effect on the final tree. Although some authors remove such problematic regions, either manually or using automatic methods, in order to improve phylogenetic performance, others prefer to keep such regions to avoid losing any information. Our aim in the present work was to examine whether phylogenetic reconstruction improves after alignment cleaning or not. Using simulated protein alignments with gaps, we tested the relative performance in diverse phylogenetic analyses of the whole alignments versus the alignments with problematic regions removed with our previously developed Gblocks program. We also tested the performance of more or less stringent conditions in the selection of blocks. Alignments constructed with different alignment methods (ClustalW, Mafft, and Probcons) were used to estimate phylogenetic trees by maximum likelihood, neighbor joining, and parsimony. We show that, in most alignment conditions, and for alignments that are not too short, removal of blocks leads to better trees. That is, despite losing some information, there is an increase in the actual phylogenetic signal. Overall, the best trees are obtained by maximum-likelihood reconstruction of alignments cleaned by Gblocks. In general, a relaxed selection of blocks is better for short alignment, whereas a stringent selection is more adequate for longer ones. Finally, we show that cleaned alignments produce better topologies although, paradoxically, with lower bootstrap. This indicates that divergent and problematic alignment regions may lead, when present, to apparently better supported although, in fact, more biased topologies.
A novel arctic-alpine lichen from Deosai National Park, Gilgit Baltistan, Pakistan
The enumeration of lichenized fungi in China
Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics
Hidden species diversity was explored in two genera of Catapyrenioid Lichens (Verrucariaceae, Ascomycota) from the deserts of China
DOI:10.3390/jof8070729
URL
[本文引用: 3]

Verrucariaceae is the third-largest lichen family with high species diversity. However, this diversity has not been well-explored in China. We carried out a wide-scale field investigation in the arid and semi-arid regions of Northwest China from 2017 to 2021. A large number of lichen groups, especially those commonly distributed in deserts, were collected. Based on molecular phylogeny using ITS and nuLSU sequences by Bayesian and maximum likelihood analyses, combining morphological characters, seven taxa of catapyrenioid lichens in Verricariaceae were found in this study, including one genus (Clavascidium) and one species (Clavascidium lacinulatum) new to China; one genus (Placidium) new to the mainland of China; and four species (Clavascidium sinense, Placidium nitidulum, Placidium nigrum, and Placidium varium) new to science. It enriched our understanding of the high species diversity in Verrucariaceae and the lichen flora of Chinese arid and semi-arid deserts.