 PDF(643 KB)
PDF(643 KB)


氨基酸影响尖孢镰孢菌古巴专化型厚垣孢子的形成
丁兆建,漆艳香,曾凡云,朱为菊,许天委,彭军,谢艺贤,张欣
菌物学报 ›› 2021, Vol. 40 ›› Issue (6) : 1413-1426.
 PDF(643 KB)
PDF(643 KB)
 PDF(643 KB)
PDF(643 KB)
氨基酸影响尖孢镰孢菌古巴专化型厚垣孢子的形成
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Amino acid involved in chlamydospore formation of Fusarium oxysporum f. sp. cubense
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}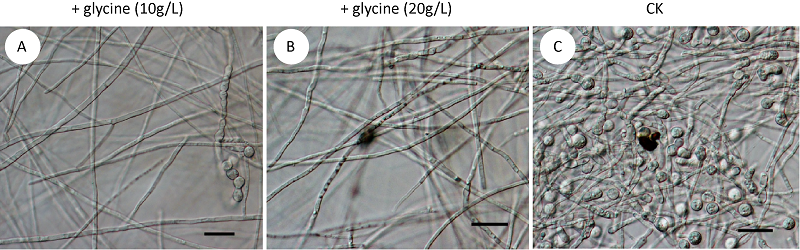
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |