INTRODUCTION
The genus Mariannaea G. Arnaud ex Samson was erected to accommodateM.elegans (Corda) Samson (type species),M.camptospora Samson and M.elegans var.punicea (Samson 1974). It is characterized by septate, branched conidiophores with hyaline, flask-shaped phialides, 1-2-celled hyaline conidia mostly forming imbricate chains or slimy heads (Samson 1974; Samson & Bigg 1988; Samuels & Seifert 1991). Sexual morphs have been linked to Nectria (Fr.) Fr. (Samuels & Seifert 1991) and Cosmospora Rabenh. (Gräfenhanet al. 2011 ). The genus was assigned to the family Nectriaceae (Samuels & Seifert 1991), which has been proved to be a monophyletic group in the family by molecular studies (Cai et al. 2010 ; Lombard et al. 2015 ; Hu et al. 2016 ). Hu et al. (2016) reviewed the genus and designated an epitype of the type species M.elegans with ex-type culture. In their study, M.elegans var. punicea Samson was raised to species rank (Hu et al. 2016 ). Mariannaea clavispora Samson & Bigg was excluded because of its cylindrical phialides, straight conidial chains and deviating phylogenetic affinity, while M.nipponica Matsush was excluded because it did not fit the generic concept of Mariannaea, and its generic placement remains uncertain (Hu et al. 2016 ). A key for 15 species was provided by Hu et al. (2016) . Later, two new species were described from saprobic habitats (Crous et al. 2019 ; Hyde et al. 2020 ).
As a part of surveys on lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region (Hyde et al. 2016 ), we have been studying lignicolous freshwater fungi in the Greater Mekong Subregion initiated in 2011 (Zhang et al. 2016 , 2017; Dong et al. 2018 , 2020; Wei et al. 2018 ; Yu et al. 2018 ; Wang et al. 2019 ; Yang et al. 2020 ). In this paper, four collections of Mariannaea were encountered from Thailand and China. To clarify the classification of them, a combined sequence dataset of LSU and ITS gene regions is analyzed and morphological characters are compared. One new species, Mariannaea submersa, is proposed.
1 MATERIALS AND METHODS
1.1 Isolation and morphology
Pieces of decaying wood were collected from small streams in Chiang Rai Province, Thailand and Yunnan Province, China following the procedures described in Kurniawati et al. (2010) . The samples were placed in ziploc plastic bags with moist sterile tissue, taken to the laboratory, and incubated at room temperature (25°C). After 1-2 weeks, specimens were examined using a stereomicroscope (SMZ-171) to locate fruiting bodies (Taylor & Hyde 2003). Photographs were taken using a Cannon EOS 600D camera attached to a Nikon ECLIPSE Ni compound microscope. The fungal structures were measured using Tarosoft (R) Image FrameWork program, and images were processed using Adobe Photoshop CS6 Extended version 13.0 software (Adobe Systems, USA). Culture isolations were made from single conidia as described byChomnunti et al. (2014) . Water agar (WA) was used for conidial germination and incubation overnight in an incubator at room temperature. Single germinating conidia were selected and transferred to a new potato dextrose agar (PDA) plate to obtain pure cultures. The colonies were checked every 3 days. Herbarium specimens are deposited in the herbarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand; and Research Institute of Resources Insects, China Academy of Forestry (IFRD). Living cultures are deposited in the Mae Fah Luang University Culture Collection (MFLUCC) and Kunming Institute of Botany, Chinese Academy of Sciences Culture Collection (KUMCC). Facesoffungi and Index Fungorum numbers are registered as described by Jayasiri et al. (2015) .
1.2 DNA extraction, PCR amplification, and sequencing
Cultures were grown on PDA at 25°C until sufficient mycelia were obtained, and a Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology Co., Ltd., Hangzhou, China) was used to extract total genomic DNA from the fresh mycelia following the manufacturer’s instructions. DNA amplification was performed using the polymerase chain reaction (PCR). Fragments of two loci, LSU and ITS, were used for phylogenetic analyses, and the following primer pairs, LROR/LR5, and ITS5/ITS4 were used for amplification and sequencing (Vilgalys & Hester 1990; White et al. 1990 ). Amplifications were performed in a 25μL reaction containing 9.5μL ddH2O, 12.5μL 2× PCR Master Mix, 1μL of DNA template, and 1μL of each primer (10μmol/L). The PCR thermal cycles for amplification of the gene regions were as inSu et al. (2015) . The PCR products were examined on 1.0% agarose electrophoresis gel stained with ethidium bromide. Sequencing reactions were conducted by Shanghai Sangon Biological Engineering Technology and Services Co., Shanghai, China.
1.3 Sequence alignment and phylogenetic analyses
Sequences were edited with BioEdit. The newly generated sequences together with other sequences obtained from GenBank were initially aligned using MAFFTv.7 (
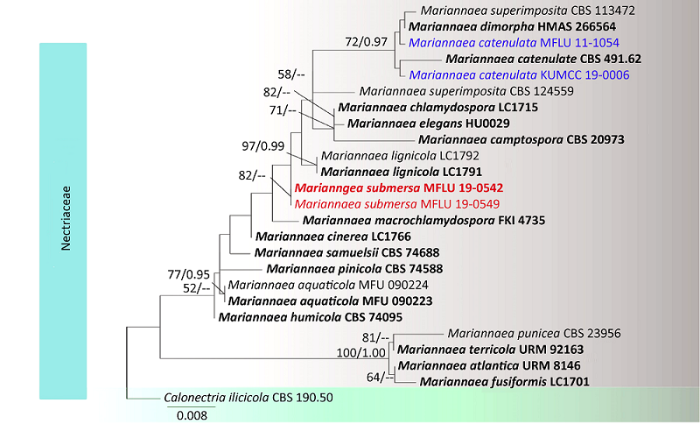
The Maximum Likelihood analysis was performed using RAxMLGUI v. 1.3 (Silvestro & Michalak 2011). The optimal ML tree search was conducted with 1 000 separate runs, using the default algorithm of the program from a random starting tree for each run. The final tree was selected among suboptimal trees from each run by comparing likelihood scores under the GTR + GAMMA substitution model. Maximum Likelihood bootstrap values equal to or greater than 50% is given as the first set of numbers above the nodes (first value, Fig. 1).
Fig. 1
Fig. 1
RAxML tree for Mariannaea species based on analyses of LSU and ITS sequence data.
The outgroup is Calonectria ilicicola (Nectriaceae). Bootstrap support values for maximum likelihood (ML, first value) equal or greater than 50% and Bayesian Posterior Probabilities from MCMC analyses (BYPP, second value) equal or higher than 0.95 are given above the nodes. Newly generated sequences are indicated in blue and new species are in red. Ex-type is in bold.
Bayesian analysis was conducted with MrBayes v.3.0b4 (Ronquist & Huelsenbeck 2003) and Posterior Probabilities (BYPP) was evaluated by Markov chain Monte Carlo (BMCMC) sampling (Rannala & Yang 1996; Zhaxybayeva & Gogarten 2002). The best-fit model is GTR+I+G for LSU and SYM+I+G for ITS. Six simultaneous Markov chains were run for 5 000 000 generations and trees were sampled every 1 000th generation and 5 000 trees were obtained. The first 1 250 trees representingthe burn-in phase of the analyses were discarded while the remaining 3 750 trees were used for calculating posterior probabilities in the majority rule consensus tree (critical value for the topological convergence diagnostic is 0.01). BYPP equal to or greater than 0.5 is given above each node (second value,Fig. 1).
Table 1 Isolates used in this study and their GenBank accession numbers
| Species | Collection/Isolate number | GenBank accession number | |
|---|---|---|---|
| LSU | ITS | ||
| Calonectria ilicicola | CBS 190.50 | GQ280727 | GQ280605 |
| Mariannaea aquaticola | MFU 090224 | GQ153835 | GQ153836 |
| Mariannaea aquaticola | MFU 090223 | GQ153833 | GQ153834 |
| Mariannaea atlantica | URM 8146 | MN151398 | MN151372 |
| Mariannaea camptospora | CBS 20973 | - | AY624202 |
| Mariannaea catenulata | CBS 491.62 | KM231617 | KM231752 |
| Mariannaea catenulata | MFLU 11-1054 | MT496750 | MT496742 |
| Mariannaea catenulata | KUMCC 19-0006 | MT510191 | MT510194 |
| Mariannaea chlamydospora | LC1715 | KX986141 | KX986134 |
| Mariannaea cinerea | LC 1766 | KX986142 | KX986135 |
| Mariannaea dimorpha | HMAS 266564 | KJ002443 | KF767353 |
| Mariannaea elegans | HU0029 | KX986139 | KX986132 |
| Mariannaea fusiformis | LC 1701 | KX986140 | KX986133 |
| Mariannaea humicola | CBS 74095 | KM231619 | KM231755 |
| Mariannaea lignicola | LC 1791 | KX986143 | KX986136 |
| Mariannaea lignicola | LC 1792 | KX986144 | KX986137 |
| Mariannaea macrochlamydospoa | FKI 4735 | AB855782 | AB855777 |
| Mariannaea pinicola | CBS 74588 | AY554242 | KM231754 |
| Mariannaea punicea | CBS 239.56 | JF415981 | AY624201 |
| Mariannaea samuelsii | CBS 74688 | KM231621 | KM231757 |
| Mariannaea submersa | MFLU 19-0542 | MT496751 | MT496743 |
| Mariannaea submersa | MFLU 19-0549 | MT496752 | MT496744 |
| Mariannaea superimposita | CBS 113472 | AB855785 | AB855780 |
| Mariannaea superimposita | CBS 124559 | AB855786 | AB85578 |
| Mariannaea terricola | URM 92163 | MK101012 | MK101011 |
Notes: Ex-type strains are in bold; newly generated sequences are underlined.
2 RESULTS
2.1 Phylogeny study
The combined dataset of LSU and ITS sequences comprised 25 strains including four new strains and one outgroup taxon Calonectria ilicicolaBoedijn & Reitsma, with an alignment length of 1 358 characters. The best scoring RAxML tree was selected to represent the relationships among taxa, in which a final likelihood value of -3073.939279 is presented ( Fig. 1).
2.2 Taxonomy
Fig. 2
Fig. 2
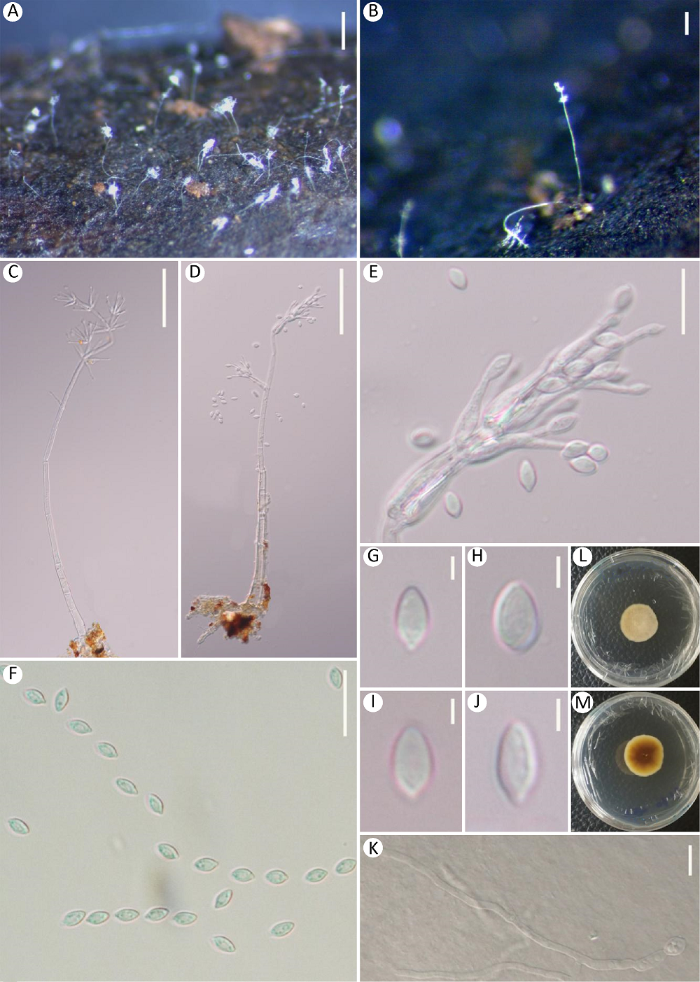
Mariannaea catenulata(MFLU 11-1054).
A: Colonies on submerged wood; B, C: Conidiophores; D-G: Upper parts of conidiophores, showing phialides; H-K: Conidia; L: Colony on PDA (from front); M: Colony on PDA (from reverse). Scale bars: A=50μm; B, C=20μm; D-K=10μm.
Fig. 3
Fig. 3
Mariannaea catenulata (KUMCC 19-0006).
A, B: Colonies on submerged wood; C, D: Conidiophores. E-G: Upper parts of conidiophores, showing phialides; H-K: Conidia; L: Germinating conidium; M: Colony on PDA (from front); N: Colony on PDA (from reverse). Scale bars: A, B=50μm; C, D=20μm; H-L=5μm.
Index Fungorum IF827817
Saprobic on decaying, submerged wood in freshwater. Asexual morph: Colonies on the substratum superficial, effuse, scattered or gregarious, hairy, white, with glistening conidia at tip of conidiophores. Mycelium partly immersed, composed of septate, branched, smooth, hyaline hyphae. Conidiophores macronematous, mononematous, erect, flexuous, straight or slightly curved, cylindrical, septate, hyaline when young, becoming medium brown at base and gradually paler towards apex in age, hyaline at apex, smooth-walled, 150-180×2-5μm ($\bar{x}$=160×3.6μm, n=10), uneven in width, robust at the base, 4-5μm wide, slender at apex 1-3μm wide, bearing short branches in the upper part with 3 phialides in verticils. Phialides flask-shaped, hyaline, slightly swollen at base, tapering at apex, not flared but with a periclinal thickening at apex, hyaline, 20-25×2-4μm ($\bar{x}$=24×2.5μm, n=20). Conidia oblong to fusiform, straight, (0-)1-septate, slightly constricted at septum, hyaline, smooth, thin-walled, with protuberant ends, acuminate at apex, narrow, truncate at base, 11-15×3-5μm ($\bar{x}$=12.6×3.6μm, n=20). Sexual morph: SeeSamuels 1985.
Culture characteristics: On PDA colony circular, reaching 15mm in 10d at 25°C, pale brown to light reddish brown at a periphery, light brown at the middle from above, brown to reddish brown from below, densely hairy in the middle, rough, dry, edge smooth.
Material examined: THAILAND, Chiang Rai, Khun Korn Waterfall, on decaying wood submerged in a stream, 26 February 2011, Huang Zhang, e49 (MFLU 11-1054), living culture MFLUCC 11-0293; ibid MFLU 11-1040. CHINA, Yunnan, Puzhehei, on decaying wood submerged in a stream, 12 May 2018, Hao Yang, P11 (IFRD500-003), living culture KUMCC 19-0006.
Distribution: China (this study), Ecuador (Samuels 1985), Jamaica (Samuels 1985), Thailand (this study), Venezuela (Samuels 1985).
Notes: In phylogenetic analysis, the two new collections aligned in the clade withMariannaea catenulata, M.dimorpha Z.Q. Zeng & W.Y. Zhuang and M.superimposita(Matsush.) Samuels, all of which are in common with 2-celled conidia. Within this clade, M.dimorpha differs in producing dimorphic conidia (microconidia and macroconidia) (Zeng & Zhuang 2014). Mariannaea superimposita differs in its conidia forming non-persistent, imbricate chains (Samuels 1989). The two new collections morphologically fit well with the characters ofM. catenulata except for shorter conidiophores (150-180μm vs. 350-450μm) (Samuels 1985). In terms of pairwise sequence divergence, MFLUCC 11-1054 differs from ex-type strains of M.catenulata CBS 491.62 by 8 and 2 nucleotide substitution, respectively in LSU and ITS regions (Table 2), which are within the generally accepted norm that 1.5% nucleotide differences in ITS regions should be one species (Luo et al. 2019 ; Dong et al. 2020 ; Yang et al. 2020 ). Mariannaeacatenulata has earlier been reported on bark from Ecuador, Jamaica and Venezuela (Samuels 1985). This is the first report from freshwater habitat and first report from Asia.
Table 2 LSU and ITS gene comparisons of Mariannaea catenulata: CBS 491.62 (type species), MFLUCC 11-1054 and KUMCC 19-0006
| Mariannaea catenulata | LSU region | ITS region | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 146 | 364 | 367 | 369 | 392 | 393 | 396 | 415 | 484 | 529 | 148 | 282 | ||
| CBS 491.62 (type) | C | T | G | C | G | - | T | T | T | T | C | T | |
| MFLUCC 11-1054 | C | A | T | G | T | C | C | T | C | C | T | C | |
| KUMCC 19-0006 | T | A | T | G | T | C | C | C | C | T | C | C | |
Mariannaea submersaH. Yang & H. Zhang, sp. nov.Fig. 4
Fig. 4
Fig. 4
Mariannaea submersa (holotype, MFLU 19-0542).
A, B: Colonies on wood; C, D: Conidiophores with conidia; E: Conidiogenous cells with conidia; F-G: Conidia; K: Germinating conidium; L: Colony on PDA (from front); M: Colony on PDA (from reverse). Scale bars: A=500μm; B=100μm; C, D=50μm; E, F, K=10μm; G-J=2μm
Etymology: The epithet refers to the submerged substrate where the fungus was first found.
Index Fungorum IF557759
Saprobic on submerged decaying wood. Asexual morph: Colonies on the substratum superficial, effuse, scattered or gregarious,hairy, white, with visible conidia on the top of conidiophores. Mycelium immersed, composed of septate, branched, smooth, hyaline hyphae. Conidiophores macronematous, mononematous, hyaline, erect, flexuous, straight or slightly curved, cylindrical, septate, hyaline, smooth- walled, 268-396μm long, 2.6-3.4μm wide at the base, 1.2-1.5μm wide at the top, bearing short branches in the upper part with 3-6 phialides in verticils. Phialides slightly swollen at base, tapering towards apex, flask-like to awl-shaped, not flared at apex, hyaline, 14-25.5×2-4μm ($\bar{x}$=19×3μm, n=25). Conidia broadly fusiform to ovoid, aseptate, hyaline with periclinal thickening at tip, smooth, thin-walled, narrowly truncated at base, acuminated with a small protuberance at the tip, produced in imbricate chains, 3.6-6.2× 2.3-3.6μm ($\bar{x}$=5.4×2.8μm, n=40). Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24h and germ tubes produced from ends or the base. Colonies on PDA fast-growing, attaining 3.2-3.5mm diameter in 15d at 25°C in the condition of 12h-dark and 12h-light, with dense, white to pale yellowish mycelium on the surface, reverse dark brown at the center, with white regular margin.
Material examined: THAILAND, Mukdahan Province, small river of Nong Bo Na Kae, on dead submerged decaying wood of unidentified plants, 13 December 2018, Hao Yang, t32 (MFLU 19-0542, holotype), ex-type living culture=MFLUCC 19-0333; ibid t42 (MFLU 19-0549, isotype), living culture=MFLUCC 19-0339 (Additional SSU sequences GenBank MT496748 for MFLU 19-0542 and MT496749 for MFLU 19-0549).
Distribution: Thailand (this study).
Notes: In phylogenetic analyses, the two strains of Mariannaea submersa form a distinct lineage in Mariannaea (Fig. 1). Morphologically, M.submersa fits the characters of Mariannaea in having hyaline, septate, branched conidiophores with verticillately arranged phialides in each node, and hyaline, aseptate conidia. Besides, the conidial chain, which is an important generic character in Mariannaea, is also observed (Fig. 4F).
3 DISCUSSION
LSU and ITS sequences are important markers for identification of Mariannaea species (Crous et al. 2019 ; Hyde et al. 2020 ). In our study, the phylogenetic relationships of species ofMariannaea are inferred on the base of LSU and ITS sequences, which also shows that Mariannaea submersa is a distinct lineage. The imbricate conidial chain was treated as an important character to differentiate Mariannaea from the similar genera such as Paecilomyces and Verticillium (Hu et al. 2016 ), which is obvious on Fig. 3A and Fig. 4F.
Species of Mariannaea were mostly reported earlier from terrestrial habitats (Samson 1974; Samson & Bigg 1988; Matsushima 1989; Samuels 1989; Samuels & Seifert 1991; Tokumasu et al. 1994 ; Gräfenhanet al. 2011 ; Zeng & Zhuang 2014; Lombard et al. 2015 ; Nonaka et al. 2015 ). Cai et al. (2010) reported the first freshwater species, Mariannaea aquaticola, and they suspected that many isolates from freshwater habitats identified as “ Verticillium” in their previous studies might belong toMariannaea. As follow-up work to examine those “Verticillium- like” species, Huet al. 2016 described four new species from freshwater, including M.chlamydosporaD.M. Hu & L. Cai, M.cinereaD.M. Hu & L. Cai, M.fusiformisD.M. Hu & L. Cai, and M.lignicolaD.M. Hu & L. Cai. In this study, we introduce a new species, M.submersa, and a new habitat and geographical report, i.e. M.catenulata from freshwater of Thailand. To date, 18 species are currently accepted in Mariannaea. All species have hyaline conidiophores except for M.catenulata and M.lignicola (Huet al. 2016 ; and this study). Most species possess aseptate conidia, while M.catenulata, M.dimorpha andM.superimposita have 1-septate conidia, and cluster in one clade in our phylogenetic tree (Fig. 1). This study further proves a high diversity of Mariannaea species in freshwater habitats.
Acknowledgments: G.N. Wang would like to thank Rekhani Hansika Perera and Niamali Indeewari de Silva for their valuable suggestions and help.
参考文献
Morphological and molecular characterization of Mariannaea aquaticola sp. nov. collected from freshwater habitats
DOI:10.1007/s11557-009-0641-1 URL [本文引用: 1]
The sooty moulds
DOI:10.1007/s13225-014-0278-5 URL [本文引用: 1]
Fungal Planet description sheets: 868-950
DOI:10.3767/persoonia.2019.42.11 URL [本文引用: 2]
Introducing Aculeata aquatica gen. et sp. nov., Minimelanolocus thailandensis sp. nov. and Thysanorea aquatica sp. nov. (Herpotrichiellaceae, Chaetothyriales) from freshwater in northern Thailand
DOI:10.1007/s11557-018-1389-2 URL [本文引用: 1]
Pseudobactrodesmium (Dactylosporaceae, Eurotiomycetes, Fungi) a novel lignicolous genus
DOI:10.3389/fmicb.2020.00456
PMID:32300334
[本文引用: 2]

During our ongoing surveys of fungi on submerged wood in the Greater Mekong Subregion, we collected two new species similar to. gen. nov. is introduced to accommodate the new species,, and is transferred to this genus. Fasciculate conidiophores, enteroblastic conidiogenous cells and subulate to fusiform, phragmoseptate conidia with a tapering apical cell and sheath characterize the genus. has longer conidia than. The placement of in (Eurotiomycetes) is a novel finding based on analyses of combined LSU, SSU, ITS and RPB2 sequence data. Our study reveals that is likely to be a speciose genus with different species in streams around the world. Copyright © 2020 Dong, Hyde, Doilom, Yu, Bhat, Jeewon, Boonmee, Wang, Nalumpang and Zhang.
An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella
DOI:10.3114/sim.2011.68.04 URL [本文引用: 2]
Phylogenetic assessment and taxonomic revision of Mariannaea
DOI:10.1007/s11557-016-1245-1 URL [本文引用: 8]
Fungal diversity notes 1151-1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa
DOI:10.1007/s13225-020-00439-5 URL [本文引用: 3]
Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function?
DOI:10.1016/j.funeco.2015.07.002 URL [本文引用: 1]
The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts
DOI:10.1007/s13225-015-0351-8 URL [本文引用: 1]
MAFFT multiple sequence alignment software version 7: improvements in performance and usability
DOI:10.1093/molbev/mst010 URL [本文引用: 1]
Diversity of freshwater ascomycetes in freshwater bodies at Amphoe Mae Chan, Chiang Rai
Generic concepts in Nectriaceae
DOI:10.1016/j.simyco.2014.12.002 URL [本文引用: 2]
Freshwater Sordariomycetes
DOI:10.1007/s13225-019-00438-1 URL [本文引用: 1]
Matsushima mycological memoirs No. 6. Matsushima fungus collect
Mariannaea macrochlamydospora, a new hyphomycete (Nectriaceae) from soil in the Bonin Islands, Japan
DOI:10.1016/j.myc.2014.02.001 URL [本文引用: 1]
Tree View: an application to display phylogenetic trees on personal computers
Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference
DOI:10.1007/BF02338839 URL [本文引用: 1]
MrBayes 3: Bayesian phylogenetic inference under mixed models
MrBayes 3 performs Bayesian phylogenetic analysis combining information from different data partitions or subsets evolving under different stochastic evolutionary models. This allows the user to analyze heterogeneous data sets consisting of different data types-e.g. morphological, nucleotide, and protein-and to explore a wide variety of structured models mixing partition-unique and shared parameters. The program employs MPI to parallelize Metropolis coupling on Macintosh or UNIX clusters.
A new species of Mariannaea from California
DOI:10.1080/00275514.1988.12025512 URL [本文引用: 2]
Four new species of Nectria and their Chaetopsina anamorphs
Nectria and penicillifer
DOI:10.1080/00275514.1989.12025758 URL [本文引用: 2]
Two new species of Nectria with Stilbella and Mariannaea anamorphs
raxmlGUI: a graphical front-end for RAxML
Hyphomycetes from aquatic habitats in Southern China: species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae)
DOI:10.11646/phytotaxa.226.3 URL [本文引用: 1]
Microfungi of tropical and temperate palms
Fungal succession on pine needles in Germany
DOI:10.1007/BF02268525 URL [本文引用: 1]
Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA froms everal Cryptococcus species
DOI:10.1128/jb.172.8.4238-4246.1990 URL [本文引用: 1]
Freshwater hyphomycetes in Eurotiomycetes: a new species of Minimelanolocus and a new collection of Thysanorea papuana (Herpotrichiellaceae)
DOI:10.1007/s11557-019-01473-7 URL [本文引用: 1]
Introducing Dictyochaeta aquatica sp. nov. and two new species of Chloridium (Chaetosphaeriaceae, Sordariomycetes) from aquatic habitats
DOI:10.11646/phytotaxa.362.2 URL
Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds.) PCR protocols: a guide to methods and applications
Four freshwater dematiaceous hyphomycetes in Sordariomycetes with two new species of Parafuscosporella
DOI:10.11646/phytotaxa.441.1 URL [本文引用: 2]
Cylindroconidiis aquaticus gen. et sp. nov., a new lineage of aquatic hyphomycetes in Sclerococcaceae (Eurotiornycetes)
DOI:10.11646/phytotaxa.372.1 URL [本文引用: 1]
A new holomorphic species of Mariannaea and epitypification of M.samuels
Molecular data shows Didymella aptrootii is a new genus in Bambusicolaceae
DOI:10.11646/phytotaxa.247.2 URL [本文引用: 1]
Towards a natural classification of Annulatascaceae-like taxa: introducing Atractosporales ord. nov. and six new families
DOI:10.1007/s13225-017-0387-z URL [本文引用: 1]
Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses
Horizontal gene transfer (HGT) played an important role in shaping microbial genomes. In addition to genes under sporadic selection, HGT also affects housekeeping genes and those involved in information processing, even ribosomal RNA encoding genes. Here we describe tools that provide an assessment and graphic illustration of the mosaic nature of microbial genomes. We adapted the Maximum Likelihood (ML) mapping to the analyses of all detected quartets of orthologous genes found in four genomes. We have automated the assembly and analyses of these quartets of orthologs given the selection of four genomes. We compared the ML-mapping approach to more rigorous Bayesian probability and Bootstrap mapping techniques. The latter two approaches appear to be more conservative than the ML-mapping approach, but qualitatively all three approaches give equivalent results. All three tools were tested on mitochondrial genomes, which presumably were inherited as a single linkage group. In some instances of interphylum relationships we find nearly equal numbers of quartets strongly supporting the three possible topologies. In contrast, our analyses of genome quartets containing the cyanobacterium Synechocystis sp. indicate that a large part of the cyanobacterial genome is related to that of low GC Gram positives. Other groups that had been suggested as sister groups to the cyanobacteria contain many fewer genes that group with the Synechocystis orthologs. Interdomain comparisons of genome quartets containing the archaeon Halobacterium sp. revealed that Halobacterium sp. shares more genes with Bacteria that live in the same environment than with Bacteria that are more closely related based on rRNA phylogeny. Many of these genes encode proteins involved in substrate transport and metabolism and in information storage and processing. The performed analyses demonstrate that relationships among prokaryotes cannot be accurately depicted by or inferred from the tree-like evolution of a core of rarely transferred genes; rather prokaryotic genomes are mosaics in which different parts have different evolutionary histories. Probability mapping is a valuable tool to explore the mosaic nature of genomes.