羌塘自然保护区是我国面积最大的国家级自然保护区,主要保护对象为保存完整的、独特的高寒生态系统及多种大型有蹄类动物 (Zhang et al. 2013)。生活在该区域的珍稀野生动物主要有藏羚羊Pantholops hodgsoni、藏野驴 Equus kiang和野牦牛Bos grunniens等青藏高原特有的国家级保护动物(Zhang & Jiang 2006;Chai et al. 2020)。近年来,由于野生动物栖息地遭到破坏,导致野生动物的种群数量逐渐减少(Harris et al. 1999)。因此,国内外越来越多的科研人开始关注和研究野生动物,但对西藏野生动物肠道微生物的研究相对较少。
动物的肠道微生物是一个庞大的、动态的系统。这些微生物依存于动物的肠道中,同时协助寄主完成多种生理生化功能(Wei et al. 2007;Yatsunenko et al. 2012)。肠道微生物不仅能够分泌多种物质,具有屏障病原体的功能,还可以影响肠道的发育,维持肠道的免疫系统,辅助肠道对各种营养物质进行消化和吸收(Macfarlane & Macfarlane 2007;Kim et al. 2012;Looft et al. 2012)。动物肠道中大部分微生物是细菌,但也存在一部分真菌。动物真菌的种类和数量受到宿主的饲粮类型、生活环境、品种以及生理阶段的影响(Liggenstoffer et al. 2010;Kittelmann et al. 2012;Sirohi et al. 2013)。真菌是动物胃肠道中主要的功能性厌氧微生物,具有强大的粗纤维降解能力,其能通过丰富的假根系统侵入植物的细胞壁中,可以产生纤维素酶、木质素酶和半纤维素酶等多种可以降解植物细胞壁的酶类,以分解纤维素等难于降解的物质,在提高草食类动物对粗饲料的利用率中具有重要意义(Joblin et al. 1989;Cheng et al. 2018)。
已有研究表明,动物的消化道组织结构不同会导致其肠道微生物不同。在对单胃草食动物和反刍动物肠道细菌菌群结构的研究中得到,厚壁菌门Firmicutes和拟杆菌门Bacteroidetes均为两类动物肠道菌群中最大的门,但反刍动物厚壁菌门的丰度显著高于单胃草食动物,且单胃草食动物拥有反刍动物没有的优势菌门(O’Donnell et al. 2017),而对两类动物肠道真菌的对比研究较少。因此本研究采用ITS1区域测序方法对羌塘保护区中的藏羚羊和藏野驴的肠道真菌群落结构进行研究,为今后功能性优势菌株筛选提供理论依据,也为研究野生动物的消化机制做出贡献。
1 材料与方法
1.1 试验动物和样品采集
本研究以西藏羌塘自然保护区的藏羚羊和藏野驴为研究对象,该区域的平均海拔在5 000m以上;气候寒冷而干燥,空气稀薄,自然环境严酷。在保护区内四处地点(32°73ʹN,88°91ʹE,海拔4 969m;32°64ʹN,88°92ʹE,海拔4 878m;32°82ʹN,88°87ʹE,海拔4 952m;32°68ʹN,88°93ʹE,海拔4 903m)分别发现藏羚羊和藏野驴各2群(藏羚羊约10只、藏野驴约9只)。采用望远镜追踪观察野生动物的排粪,当动物离开后随机挑拣两种野生动物的新鲜粪样各5份,用灭菌牙签将粪便外层拨开,挑取里面没有污染的新鲜粪便装入20mL的冻存管中,并在试管表面进行标记:A组(A1、A2、A3、A4、A5)为5份藏羚羊粪样、B组(B1、B2、B3、B4、B5)为5份藏野驴粪样。所有粪样均放入车载冰箱中(-16℃)运回实验室进行粪便真菌检测。
1.2 粪便总DNA的提取和真菌ITS1区的扩增
使用QIAGEN公司的QIAamp Fast DNA Stool Mini试剂盒提取藏羚羊和藏野驴粪便的总DNA。通过紫外分光光度法和琼脂糖凝胶电泳对提取的DNA进行质量检测。将提取的DNA统一稀释到20ng/μL,不足的直接使用原液。以稀释后的DNA为模板,使用引物:ITS1f(5’-TCCGTAGGTGAACCTGCGG-3’)和ITS2(5’-TCCTCCGCTTATTGATATGC-3’)对真菌ITS1区进行PCR扩增。扩增体系(25μL)为:5×Reaction Buffer 5μL,5×GC缓冲液5μL,dNTP(2.5mmol/L)2μL,前引物(10μmol/L)1μL,后引物(10μmol/L)1μL,DNA模板2μL,ddH2O 8.75μL,Q5 DNA聚合酶0.25μL。扩增参数为:98℃预变性2min;98℃变性15s,55℃退火30s,72℃延伸30s,30个循环;72℃延伸5min;4℃保存。
1.3 扩增产物回收纯化和荧光定量
PCR扩增产物通过2%琼脂糖凝胶电泳进行检测,采用AXYGEN公司的凝胶回收试剂盒对目标片段进行切胶回收,参照电泳初步定量结果,利用Quant-iT PicoGreen dsDNA Assay Kit对扩增回收产物进行荧光定量。根据荧光定量结果,按照每个样本的测序量需求,对各样本按相应比例进行混合。
1.4 测序文库制备和高通量测序
采用Illumina公司的TruSeq Nano DNA LT Library Prep Kit制备测序文库,采用Agilent High Sensitivity DNA Kit对文库在Agilent Bioanalyzer上进行质检。采用Quant-iT PicoGreen dsDNA Assay Kit在Promega QuantiFluor荧光定量系统上对文库进行定量。将合格的各上机测序文库(Index序列不可重复)梯度稀释后,根据所需测序量按相应比例混合。通过琼脂糖凝胶电泳(2%),对文库做最终的选择与纯化。利用Illumina MiSeq设备进行双端测序(上海派森诺生物科技股份有限公司)。
1.5 生物信息学和数据分析
本研究采用Illumina MiSeq平台对5份藏羚羊粪便和5份藏野驴粪便真菌的ITS1区域进行双端测序,对所获序列逐一作质量筛查,按照碱基平均测序准确率≥99%,序列长度≥150bp,且不允许存在模糊碱基的要求,剔除疑问序列。使用聚类程序VSEARCH(1.9.6)对获得的序列进行聚类,并以97%的序列相似性对OTU(operational taxonomic unit)进行归并划分(Bokulich et al. 2013),通过与UNITE数据库的序列相比(Koljalg et al. 2013),对OTU的代表序列进行分类地位鉴定,获取每个OTU所对应的分类学信息,将低于全体样本测序总量0.001%的OTU去除,并将去除了稀有OTU的多度矩阵用于后续分析;利用4种度量指数(Chao1、ACE、Shannon和Simpson)来计算真菌菌群的丰富度和均匀度;通过主成分分析(principal component analysis,PCA)考察不同样本之间群落结构的相似性(Ramette 2007);使用IBM SPSS Statistics软件对试验数据进行单因素方差分析。
2 结果与分析
2.1 粪便真菌测序结果和各分类水平所获菌种数
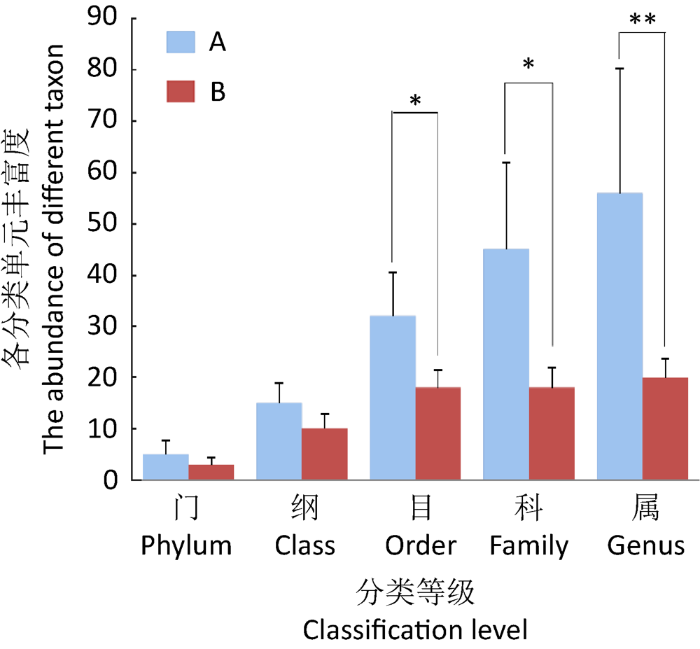
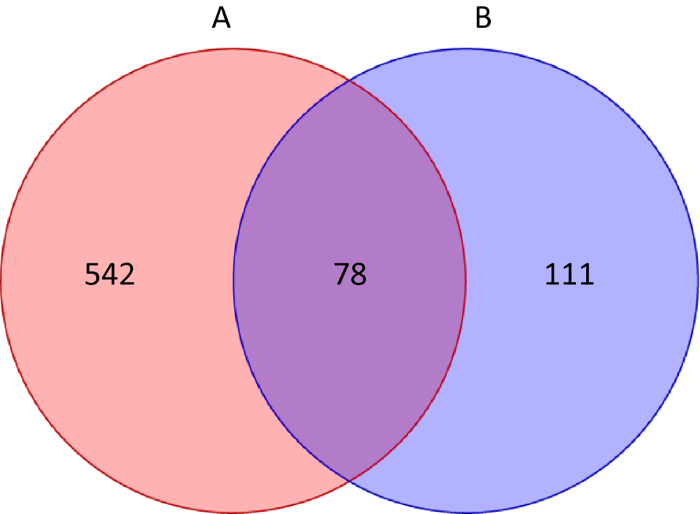
本研究分别从藏羚羊和藏野驴中获得168 822条和194 063条有效序列,全部样本中序列的长度均分布于150-350bp之间。对每个样品可以鉴定为真菌的序列数进行抽平处理,按10个样品中最低样品的序列数34 214为依据,从每个样本中分别随机抽取一定数量的序列,使10个样品的序列数均达到34 214个,从而预测各样本在该测序深度下,所能观测到的OTUs及其相对丰度。藏羚羊和藏野驴各粪便样品在门、纲、目、科、属5种分类水平下所获得的OTUs数量见图1。藏羚羊中共鉴定出5个门、15个纲、32个目、45个科和56个属;藏野驴中共鉴定出3个门、10个纲、18个目、18个科和20个属。由各分类水平的真菌类群数统计结果可知,反刍动物(藏羚羊)粪便真菌在各分类水平的类群统计数均高于单胃草食动物(藏野驴)。由Venn图可知(图2),藏羚羊和藏野驴粪便中共计获得731个OTUs,藏羚羊共有620个OTUs、藏野驴共有189个OTUs,藏羚羊和藏野驴的独有OTUs分别为542和111种,而共有OTUs为78种,独有OTUs数量均多于共有OTUs,表明藏羚羊和藏野驴粪便真菌菌群结构差异较大。
图1
图1
藏羚羊与藏野驴各分类等级的真菌类群数目
同一分类水平下,藏羚羊和藏野驴之间的差异显著性用星号表示,*P<0.05,**P<0.01
Fig. 1
Number of fungal taxa in feces of Tibetan antelope (A) and Tibetan wild ass (B) at different taxonomic level.
Asterisks represent significant differences between Tibetan antelope and Tibetan wild ass at the same taxonomic level, *P<0.05, **P<0.01.
图2
图2
藏羚羊和藏野驴粪便中真菌OTUs的Venn图
Fig. 2
Venn analysis of fecal fungal OTUs of Tibetan antelopes (A) and Tibetan wild asses (B).
2.2 藏羚羊和藏野驴粪便真菌菌群多样性分析
表1 藏羚羊和藏野驴粪便中真菌菌群多样性指数
Table 1
| 分组 Group | Simpson指数 Simpson index | Chao1指数 Chao1 index | ACE指数 ACE index | Shannon指数 Shannon index | OTUs数 OTUs number |
|---|---|---|---|---|---|
| 藏羚羊 Tibetan antelope | 0.88±0.09a | 188.30±112.72a | 188.99±112.33a | 4.89±1.02a | 153.00±81.26a |
| 藏野驴 Tibetan wild ass | 0.27±0.12b | 77.00±15.63a | 78.06±16.71a | 0.97±0.40b | 73.60±13.72a |
注:同列数字上的不同字母表示差异显著(P<0.05)
Note: The different letters indicate significant difference (P<0.05).
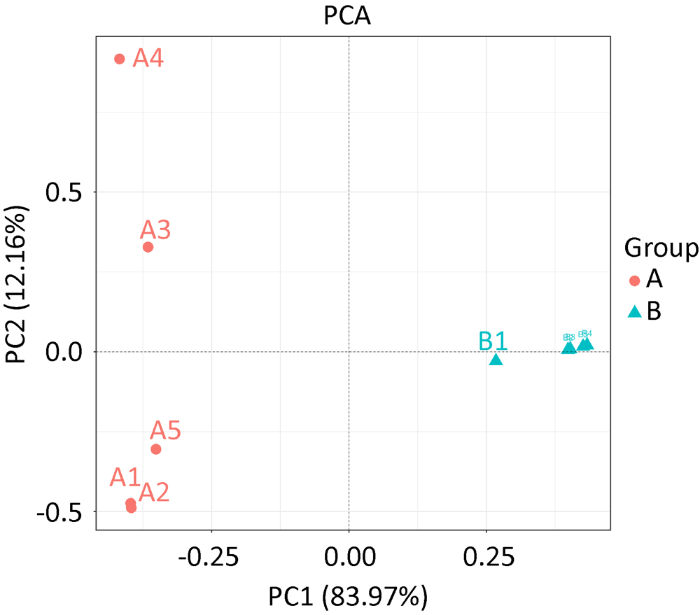
图3
图3
藏羚羊和藏野驴粪便样品真菌群落PCA图
Fig. 3
PCA analysis of fecal fungus community of Tibetan antelope and Tibetan wild ass.
2.3 藏羚羊和藏野驴粪便真菌菌群组成分析
在门分类水平上,藏羚羊和藏野驴粪便样品中共鉴定出5个真菌门,其中子囊菌门和担子菌门分别占总真菌序列数的82.70%和13.17%,4.13%为其他菌门和未鉴定的真菌(表2)。藏羚羊粪便中子囊菌门的相对多度低于藏野驴,而担子菌门的相对多度高于藏野驴,但差异均不显著(P>0.05)。在纲和目分类水平上,藏羚羊和藏野驴粪便样品中共鉴定出15个纲和32个目,其中有5个纲和6个目的相对多度大于1.0%(表2)。与藏野驴相比,藏羚羊粪便中锤舌菌纲Leotiomycetes、银耳菌纲Tremellomycetes、寡囊盘菌目Thelebolales和线黑粉菌目Filobasidiales的相对多度显著降低(P<0.05);而座囊菌纲Dothideomycetes、粪壳菌纲Sordariomycetes、微球黑粉菌纲Microbotryomycetes、格孢菌目Pleosporales、肉座菌目Hypocreales、煤炱菌目Capnodiales和柔膜菌目Helotiales的相对多度显著升高(P<0.05)。在科分类水平上,两种野生动物粪便样品中共鉴定出45个科(表2)。藏羚羊粪便中寡囊盘菌科Thelebolaceae和线黑粉菌科Filobasidiaceae的相对多度显著低于藏野驴(P<0.05);而小双腔菌科Didymellaceae、葡萄穗霉科Stachybotryaceae、荚孢腔菌科Sporormiaceae和虫草科Cordycipitaceae的相对多度显著高于藏野驴(P<0.05)。
表2 藏羚羊和藏野驴粪便中不同真菌分类水平的相对多度
Table 2
| 分类水平 Taxonomic level | 真菌 Fungus | 分组Group | |
|---|---|---|---|
| 藏羚羊 Tibetan antelope | 藏野驴 Tibetan wild ass | ||
| 门 Phylum | 子囊菌门 Ascomycota | 77.46±10.43a | 87.94±6.28a |
| 担子菌门 Basidiomycota | 14.34±11.34a | 12.00±6.32a | |
| 被孢霉门 Mortierellomycota | 0.06±0.02b | 0.00±0.00a | |
| 丝足虫类 Cercozoa | 0.05±0.02b | 0.01±0.00a | |
| 毛霉门 Mucoromycota | 0.04±0.03b | 0.01±0.00a | |
| 纲 Class | 锤舌菌纲 Leotiomycetes | 7.54±4.52a | 85.00±8.62b |
| 座囊菌纲 Dothideomycetes | 40.80±10.26b | 1.60±1.44a | |
| 粪壳菌纲 Sordariomycetes | 17.52±12.77b | 0.38±0.21a | |
| 银耳菌纲 Tremellomycetes | 3.46±1.89a | 11.94±6.30b | |
| 微球黑粉菌纲 Microbotryomycetes | 8.80±6.95b | 0.00±0.00a | |
| 散囊菌纲 Eurotiomycetes | 0.94±0.64b | 0.06±0.04a | |
| 伞菌纲 Agaricomycetes | 0.54±0.36b | 0.01±0.00a | |
| 酵母菌纲 Saccharomycetes | 0.46±0.39b | 0.04±0.03a | |
| 马拉色菌纲 Malasseziomycetes | 0.33±0.06b | 0.01±0.00a | |
| 茶渍纲 Lecanoromycetes | 0.21±0.15b | 0.01±0.00a | |
| 沃勒曼菌纲 Wallemiomycetes | 0.19±0.12b | 0.01±0.00a | |
| 盘菌纲 Pezizomycetes | 0.06±0.03a | 0.11±0.07a | |
| 外担菌纲 Exobasidiomycetes | 0.17±0.16b | 0.01±0.00a | |
| 黑粉菌纲 Ustilaginomycetes | 0.10±0.07b | 0.02±0.01a | |
| 目 Order | 寡囊盘菌目 Thelebolales | 3.28±2.25a | 84.54±8.52b |
| 格孢菌目 Pleosporales | 35.06±7.57b | 1.58±1.40a | |
| 锁掷酵母目 Sporidiobolales | 14.76±9.09b | 0.00±0.00a | |
| 肉座菌目 Hypocreales | 14.50±11.95b | 0.10±0.07a | |
| 线黑粉菌目 Filobasidiales | 2.24±1.97a | 11.88±6.33b | |
| 煤炱菌目 Capnodiales | 3.78±3.10b | 0.00±0.00a | |
| 柔膜菌目 Helotiales | 3.00±1.35b | 0.16±0.11a | |
| 小丛壳目 Glomerellales | 1.68±0.62b | 0.01±0.00a | |
| 丝孢酵母目 Trichosporonales | 0.74±0.29b | 0.04±0.02a | |
| 白粉菌目 Erysiphales | 0.76±0.27b | 0.01±0.00a | |
| 粪壳菌目 Sordariales | 0.72±0.30b | 0.00±0.00a | |
| 禾本目 Poales | 0.62±0.31b | 0.00±0.00a | |
| 座囊菌纲未定目 Dothideomycetes ord. Incertae sedis | 0.60±0.53b | 0.00±0.00a | |
| 散囊菌目 Eurotiales | 0.47±0.45a | 0.06±0.03a | |
| 酵母菌目 Saccharomycetales | 0.46±0.44a | 0.04±0.02a | |
| 小囊菌目 Microascales | 0.44±0.32b | 0.01±0.00a | |
| Coniochaetales | 0.20±0.09a | 0.19±0.16a | |
| 马拉色菌目 Malasseziales | 0.34±0.31a | 0.03±0.02a | |
| 刺盾炱目 Chaetothyriales | 0.15±0.02b | 0.00±0.00a | |
| 银耳菌目 Tremellales | 0.24±0.15b | 0.00±0.00a | |
| Togniniales | 0.23±0.13b | 0.01±0.00a | |
| 座囊菌目 Dothideales | 0.22±0.13b | 0.00±0.00a | |
| 科 Family | 寡囊盘菌科 Thelebolaceae | 3.28±2.25a | 84.54±8.52b |
| 锁掷酵母科 Sporidiobolaceae | 14.76±9.09b | 0.00±0.00a | |
| 线黑粉菌科 Filobasidiaceae | 2.18±1.96a | 11.88±6.33b | |
| 小双腔菌科 Didymellaceae | 13.88±10.82b | 0.00±0.00a | |
| 葡萄穗霉科 Stachybotryaceae | 11.12±10.77b | 0.00±0.00a | |
| 荚孢腔菌科 Sporormiaceae | 3.18±2.64b | 0.40±0.31a | |
| 虫草科 Cordycipitaceae | 1.92±1.05b | 0.06±0.05a | |
| 织球壳科 Plectosphaerellaceae | 1.44±0.87b | 0.01±0.00a | |
| 蛹孢假壳科 Leptosphaeriaceae | 1.62±0.96b | 0.06±0.05a | |
| 畸球腔菌科 Teratosphaeriaceae | 1.10±0.53b | 0.00±0.00a | |
| 球腔菌科 Mycosphaerellaceae | 0.99±0.37b | 0.00±0.00a | |
| 枝孢菌科 Cladosporiaceae | 0.96±0.42b | 0.00±0.00a | |
| 丛赤壳科 Nectriaceae | 0.81±0.27b | 0.00±0.00a | |
| 丝孢酵母科Trichosporonaceae | 0.74±0.29b | 0.04±0.02a | |
| 白粉菌科 Erysiphaceae | 0.76±0.36b | 0.00±0.00a | |
| 禾本科 Poaceae | 0.62±0.31b | 0.00±0.00a | |
| 座囊菌纲未定科 Dothideomycetes fam. Incertae sedis | 0.60±0.52b | 0.00±0.00a | |
| 暗球壳科Phaeosphaeriaceae | 0.54±0.27b | 0.00±0.00a | |
| 煤炱科 Capnodiaceae | 0.49±0.30b | 0.00±0.00a | |
| 晶杯菌科 Hyaloscyphaceae | 0.46±0.34b | 0.00±0.00a | |
| 生赤壳科 Bionectriaceae | 0.44±0.28b | 0.00±0.00a | |
| 曲霉科 Aspergillaceae | 0.38±0.36a | 0.05±0.03a | |
| 毛孢壳科 Coniochaetaceae | 0.19±0.03a | 0.22±0.20a | |
| 格孢腔菌科 Pleosporaceae | 0.32±0.20b | 0.00±0.00a | |
| 毛壳菌科 Chaetomiaceae | 0.31±0.15b | 0.00±0.00a | |
| Togniniaceae | 0.23±0.19b | 0.01±0.00a | |
| 黏毛菌科 Myxotrichaceae | 0.23±0.09b | 0.00±0.00a | |
| 属 Genus | 寡囊盘菌属 Thelebolus | 3.28±2.25a | 84.54±8.52b |
| 掷孢酵母属 Sporobolomyces | 14.76±9.09b | 0.00±0.00a | |
| Naganishia | 2.18±1.96a | 11.88±6.33b | |
| 亚隔孢壳属 Didymella | 13.80±10.85b | 0.00±0.00a | |
| Plenodomus | 1.33±0.61b | 0.00±0.00a | |
| 蜡蚧轮枝菌属 Lecanicillium | 1.27±0.15b | 0.04±0.02a | |
| 织球菌属 Plectosphaerella | 1.19±0.43b | 0.00±0.00a | |
| 石菌属 Lapidomyces | 1.10±0.52b | 0.00±0.00a | |
| 枝孢菌属 Cladosporium | 0.96±0.42b | 0.01±0.00a | |
| 多胞小球壳属 Sphaerulina | 0.80±0.24b | 0.00±0.00a | |
| 小粉孢属 Microidium | 0.76±0.36b | 0.01±0.00a | |
| 皮生丝孢酵母属 Cutaneotrichosporon | 0.71±0.30b | 0.04±0.02a | |
| 简青霉属 Simplicillium | 0.71±0.25b | 0.02±0.01a | |
| 漆斑菌属 Myrothecium | 0.73±0.30b | 0.00±0.00a | |
| 门座菌属 Thyrostroma | 0.60±0.52b | 0.00±0.00a | |
| 小角炱属 Antennariella | 0.49±0.26b | 0.00±0.00a | |
| 粉红螺旋聚孢霉属 Clonostachys | 0.44±0.28b | 0.00±0.00a | |
| 轮枝菌属 Verticillium | 0.41±0.22b | 0.01±0.00a | |
| Cistella | 0.42±0.36b | 0.00±0.00a | |
| 新壳多孢属 Neostagonospora | 0.37±0.24b | 0.01±0.00a | |
| 腐质霉属 Humicola | 0.25±0.16b | 0.00±0.00a | |
| 暗色枝顶孢属 Phaeoacremonium | 0.23±0.11b | 0.01±0.00a | |
| 马拉色氏霉菌属 Malassezia | 0.20±0.19b | 0.01±0.00a | |
| 镰刀菌属 Fusarium | 0.20±0.10b | 0.00±0.00a | |
| 树粉孢属 Oidiodendron | 0.18±0.13b | 0.00±0.00a | |
| 黑团孢属 Periconia | 0.18±0.08b | 0.00±0.00a | |
| 支顶孢属 Acremonium | 0.18±0.08b | 0.00±0.00a | |
| 顶囊菌属 Claussenomyces | 0.14±0.13b | 0.00±0.00a | |
| 金孢属 Chrysosporium | 0.13±0.08b | 0.00±0.00a | |
| 青霉菌属 Penicillium | 0.12±0.10b | 0.01±0.00a | |
| 盘菌属 Peziza | 0.00±0.00a | 0.11±0.09b | |
| 黑粉菌属 Ustilago | 0.03±0.02b | 0.00±0.00a | |
注:同列数字上不同字母表示差异显著(P<0.05)
Note: The different letters in the same column indicate significant difference (P<0.05).
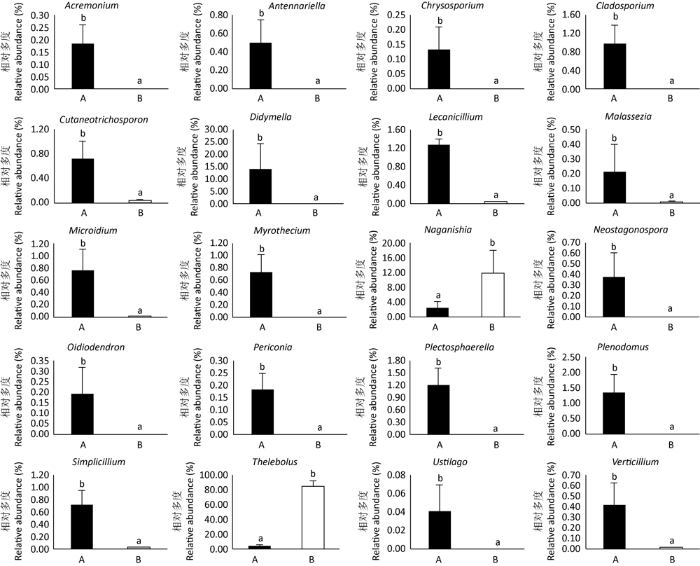
比较分析藏羚羊和藏野驴在真菌属分类水平下的差异,两种野生动物共有34属真菌的相对多度存在显著性差异(P<0.05,图4)。其中,Antennariella、枝顶孢属Acremonium、黑团孢霉属Periconia、Plectosphaerella、丰屋菌属Plenodomus、漆斑菌属Myrothecium、Oidiodendron、金孢菌属Chrysosporium、Hypomyces、Clonostachys、多胞小球壳属Sphaerulina、Thyrostroma、镰刀菌属Fusarium、Lapidomyces、Termitomyces和Cistella只在藏羚羊中存在;盘菌属Peziza和Stagonospora只在藏野驴中存在。除此之外,藏野驴粪便中寡囊盘菌属和Naganishia的相对多度显著高于藏羚羊(P<0.05);而蜡蚧轮枝菌属Lecanicillium、轮枝菌属Verticillium、Cutaneotrichosporon、亚隔孢壳属、简青霉菌属Simplicillium、Neostagonospora、枝孢菌属Cladosporium、黑粉菌属Ustilago、马拉色氏霉菌属Malassezia、微杆菌属Microidium、腐质霉菌属Humicola、Phaeoacremonium、Claussenomyces和青霉菌属Penicillium的相对多度显著低于藏羚羊(P<0.05)。
图4
图4
藏羚羊和藏野驴的真菌属的相对多度比较
Fig. 4
Relative abundance of fungal genera in feces of Tibetan antelopes and Tibetan wild asses.
3 讨论
动物的粪便真菌作为一类新型功能性微生物,可以生产酶制剂、抗真菌剂、抗细菌剂等多种产品,在工业上具有重要的潜在应用价值(Weber & Gloer 1991;Soman et al. 1999;Farouq et al. 2013)。已有研究表明,动物真菌具有强大的降解粗纤维的能力,可以有效地提高动物对粗饲料的利用率,因此研究动物粪便真菌对开发新的微生物来源具有重要意义。因此本研究对同一生活环境中的反刍动物(藏羚羊)和单胃草食动物(藏野驴)的粪便真菌进行研究,可以为阐明动物消化方式对真菌菌群结构的影响奠定基础。
研究结果表明,藏羚羊和藏野驴粪便中有多种真菌分布,在藏羚羊粪便中共鉴定到56属,而在藏野驴粪便中共鉴定到20属,且藏羚羊粪便真菌的平均物种丰富度和真菌多样性均高于藏野驴,这与之前对反刍动物和兔形目动物粪便真菌多样性的研究结果相一致(Richardson 2001)。这可能与反刍动物主要依靠瘤胃对粗饲料进行微生物消化(Jiao et al. 2013),而单胃草食动物肠道中缺少可以消化粗纤维的酶类,其主要依靠肠道内的微生物降解粗纤维(Jones 2013),且动物粪便真菌会受消化道结构的影响有关。研究得到,锤舌菌纲、座囊菌纲、粪壳菌纲、银耳菌纲和微球黑粉菌纲为两种野生动物的主要优势菌群,这与对中国云南(Hu et al. 2008)、意大利(Caretta et al. 1994)、波多黎各(Richardson 2008)、福克兰群岛(Watling & Richardson 2010)、非洲(Ebersohn & Eicker 1992)等地的动物粪便真菌的研究结果不一致,这可能是由于西藏羌塘国家自然保护区高原低氧的特殊环境,以及粪便真菌受饲养环境的影响造成。
本研究还发现,藏羚羊与藏野驴粪便真菌菌群结构具有显著差异,藏羚羊粪便中有16个属在藏野驴中没有鉴定到,而藏野驴粪便中有2个属在藏羚羊粪便中没有鉴定到。除此之外藏野驴粪便中2种真菌的丰度高于藏羚羊,而12种真菌的丰度低于藏羚羊,由此表明反刍动物与单胃草食动物所利用的功能性真菌的种类不同。在对西藏羌塘地区牧草种类的调查中发现,该地区大部分面积生长着紫花针茅等植物,紫花针茅的草质较硬,粗纤维含量高,可能是藏羚羊和藏野驴等野生动物的主要牧草。已有研究表明,Necallimastix、Anaeromyces、Orpinomyces以及Piromyces(Freelove et al. 2001;James et al. 2006)等属真菌可以通过假根对植物组织的细胞壁进行分解,进而降解粗饲料中的粗纤维等物质;Caeocomyces和Cyllamyces(Chen et al. 2007)等属真菌可以通过黏附和破坏植物组织分解粗纤维。本研究在两种野生动物中均未鉴定到以上菌属,因此推断,藏羚羊和藏野驴胃肠道中可能有其他具有降解粗纤维功能的菌种,但具体菌种,有待进一步研究。Lecanicillium是害虫防治中的一种重要的生防真菌,寄主范围十分广泛,作为生防制剂已被商品化生产(Gurulingappa et al. 2011);Simplicillium代谢产生的抗生素对金黄色葡萄球菌、枯草芽孢杆菌、藤黄微球菌等革兰氏阳性菌具有较强的抑制作用,而对大肠杆菌和鼠伤寒沙门氏菌等革兰氏阴性菌无抑制作用(Chen et al. 2008;Dong et al. 2014);研究得到,藏羚羊和藏野驴粪便中均有Lecanicillium和Simplicillium等多种有益真菌,但有益真菌在两种野生动物中的丰度明显不同,这可能说明动物的免疫机制也与动物种类有关。除此之外,本研究还在两种野生动物粪便中鉴定到黑团孢霉属、轮枝菌属、Cutaneotrichosporon、黑粉菌属、Malassezia、腐质霉菌属和青霉菌属等多种致病菌,由此推断,即使是生活于高原低氧的环境下,藏羚羊和藏野驴等野生动物在生长过程中也会滋生各种条件性致病真菌。
综上所述,本研究对藏羚羊和藏野驴粪便真菌菌群多样性进行测定和分析,既可以为探究动物消化结构对粪便真菌菌群结构的影响和挖掘功能性真菌奠定基础,又可以为进一步揭示青藏高原野生动物源真菌的分布和鉴定提供一定的理论依据和技术方法。
参考文献
Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing
DOI:10.1038/nmeth.2276 URL [本文引用: 1]
Coprophilous fungi on horse, goat and sheep dung from Lombardia (Italy)
Whole-genome resequencing provides insights into the evolution and divergence of the native domestic yaks of the Qinghai-Tibet Plateau
DOI:10.1186/s12862-020-01702-8 URL [本文引用: 1]
First report of Simplicillium lanosoniveum causing brown spot on Salvinia auriculata and S. molesta in Taiwan
DOI:10.1094/PDIS-92-11-1589C
PMID:30764451
[本文引用: 1]

Salvinia spp. are small, floating ferns that grow in long chains of two oval leaves and a root-like third leaf. S. natans (L.) All., a native floating fern distributed in paddy fields, ponds, and ditches in Taiwan, has become critically endangered. Another two exotic species, S. auriculata Aublet (eared salvinia) and S. molesta Mitchell (giant salvinia), are sold in increasing frequency at local flower markets and aquarium shops and pose a serious threat when they find their way into the natural environment. Brown spot of S. auriculata was found in a home aquarium in December 2006 in Chiayi, Taiwan. Symptoms of the disease included many, irregular, dark brown spots on both upper and lower leaf surfaces. Lesions on the upper surface of the leaves were covered with white patches of mycelia and abundant conidia. Small pieces (approximately 2 × 2 mm) of diseased leaf tissue from the margin of individual lesions were surface disinfected in 1% sodium hypochlorite solution for 1 min, rinsed in sterile water, plated on water agar, and incubated at 25°C. Six isolates of the fungus were then isolated and transferred to potato dextrose agar (PDA). Isolate Cs0701 was identified morphologically as Simplicillium lanosoniveum (van Beyma) Zare & W. Gams on the basis of morphology of asexual reproduction structures and rDNA sequence analysis (1). In culture, this fungus formed whitish-to-whitish yellow, pulvinate colonies with matted surfaces. The reverse side of cultures was yellow to light brown. Small, ovate to spherical, hyaline conidia, 2.2 to 3.0 × 1.6 to 2.0 μm (average 2.4 × 1.9 μm) were formed. To confirm the identity of the fungus, PCR amplification and DNA sequencing of the internal transcribed spacer (ITS1-5.8S-ITS2 rRNA gene cluster) was conducted on isolates Cs0701 and Cs0702. The sequence of the PCR product was compared with sequences of closely related species listed in the GenBank database. Except for a single nucleotide, the ITS sequence of both isolates (480 bp; GenBank Accession No. EU939525) was identical to the rRNA of Simplicillium lanosoniveum (GenBank Accession No. AJ292396). Koch's postulates were performed to confirm the pathogenicity of the fungus on S. auriculata and S. molesta. After 14 days of growth on PDA, a spore suspension of isolate Cs0701 (10 spores per ml) was sprayed onto approximately 5 and 10 g of healthy S. auriculata and S. molesta plants, respectively, floated in 500-ml beakers filled with 300 ml of tap water. All treatments, including controls misted with sterile water, were replicated three times. The beakers were covered with plastic bags and placed in a growth chamber maintained at 25°C with 12-h fluorescent light cycles. After 2 days, the bags were removed. Symptoms developed on all inoculated plants 4 days after inoculation. In all cases, typical brown spots were observed. Simplicillium lanosoniveum was reisolated from all surface-disinfested infected tissues. Control plants developed no symptoms. Six isolates of the fungus are being maintained at the Department of Microbiology and Immunology, National Chiayi University, Taiwan. To our knowledge, this is the first report of Simplicillium lanosoniveum causing brown spot of S. auriculata and S. molesta in Taiwan. Reference: (1) R. Zare and W. Gams. Nova Hedwigia 73:1, 2001.
Caecomyces sympodialis sp. nov., a new rumen fungus isolated from Bos indicus
DOI:10.1080/15572536.2007.11832607 URL [本文引用: 1]
The biotechnological potential of anaerobic fungi on fiber degradation and metha
DOI:10.1007/s11274-018-2539-z URL [本文引用: 1]
Identification of a symbiotic fungus from blue- green alga and its extracellular polysaccharide
DOI:10.1111/lam.12192
PMID:24261819
[本文引用: 1]

A previously unknown symbiotic fungus DT06 has been isolated from the single-celled blue-green alga Chroococcus sp. The sequences of ITS1, 5.8S rDNA and ITS2 regions of DT06 have a high similarity with that of Simplicillium (98%), which is closely related to Simplicillium lanosoniveum based on further phylogenetic analysis. However, DT06 produces unusual exocellular crystals with its conidium size twice that of S. lanosoniveum. Hence, DT06 is proposed to be a varietas of S. lanosoniveum and named as S. lanosoniveum var. Tianjinienss. Dong. (Type specimen was deposited at China General Microbiological Culture Collection Center, Number: CGMCC4460.). The striking character of DT06 is its massive production of a unique extracellular polysaccharide, which is composed of glucose and galactose and linked by 1-4 and 1-6 glycoside bonds according to UV, IR and NMR analysis. Therefore, DT06 may represent a new source of bioactive products, and also, its unusual symbiotic partnership with blue-green algae provides a model for investigating the interaction between photoautotrophic and heterotrophic micro-organisms in aquatic ecosystems.A novel fungus (Simplicillium) symbiotic with a single-celled blue-green alga Chroococcus sp. and its major primary metabolite have been isolated and identified. These findings broaden the scope of symbiotic fungi and provide a unique extracellular polysaccharide with potential applications in food industry.© 2013 The Society for Applied Microbiology.
Coprophilous fungal species composition and species diversity on various dung substrates of African game animals
Isolation and characterization of coprophilous cellulolytic fungi from Asian elephant (Elephas maximus) dung
A novel carbohydrate-binding protein is a component of the plant cell wall- degrading complex of Piromyces equi
The recycling of photosynthetically fixed carbon by the action of microbial plant cell wall hydrolases is a fundamental biological process that is integral to one of the major geochemical cycles and, in addition, has considerable industrial potential. Enzyme systems that attack the plant cell wall contain noncatalytic carbohydrate-binding modules (CBMs) that mediate attachment to this composite structure and play a pivotal role in maximizing the hydrolytic process. Anaerobic fungi that colonize herbivores are the most efficient plant cell wall degraders known, and this activity is vested in a high molecular weight complex that binds tightly to the plant cell wall. To investigate whether plant cell wall attachment is mediated by noncatalytic proteins, a cDNA library of the anaerobic fungus Piromyces equi was screened for sequences that encode noncatalytic proteins that are components of the cellulase-hemicellulase complex. A 1.6-kilobase cDNA was isolated encoding a protein of 479 amino acids with a M(r) of 52548 designated NCP1. The mature protein had a modular architecture comprising three copies of the noncatalytic dockerin module that targets anaerobic fungal proteins to the cellulase-hemicellulase complex. The two C-terminal modules of NCP1, CBM29-1 and CBM29-2, respectively, exhibit 33% sequence identity with each other but have no homologues in protein data bases. A truncated form of NCP1 comprising CBM29-1 and CBM29-2 (CBM29-1-2) and each of the two individual copies of CBM29 bind primarily to mannan, cellulose, and glucomannan, displaying the highest affinity for the latter polysaccharide. CBM29-1-2 exhibits 4-45-fold higher affinity than either CBM29-1 or CBM29-2 for the various ligands, indicating that the two modules, when covalently linked, act in synergy to bind to an array of different polysaccharides. This paper provides the first report of a CBM-containing protein from an anaerobic fungal cellulase-hemicellulase complex. The two CBMs constitute a novel CBM family designated CBM29 whose members exhibit unusually wide ligand specificity. We propose, therefore, that NCP1 plays a role in sequestering the fungal enzyme complex onto the plant cell wall.
Endophytic Lecanicillium lecanii and Beauveria bassiana reduce the survival and fecundity of Aphis gossypii following contact with conidia and secondary metabolites
DOI:10.1016/j.cropro.2010.11.017 URL [本文引用: 1]
Status and trends of Tibetan plateau mammalian fauna, Yeniugou, China
DOI:10.1016/S0006-3207(98)00046-9 URL [本文引用: 1]
The diversity of coprophilous fungi from Dahuadian and Zhongdian grasslands, Yunnan, China
A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota)
DOI:10.1080/15572536.2006.11832616 URL [本文引用: 1]
In vitro evaluation on neutral detergent fiber and cellulose digestion by post-ruminal microorganisms in goats
DOI:10.1002/jsfa.6485 URL [本文引用: 1]
Fermentation of barley straw by anaerobic rumen bacteria and fungi in axenic culture and in co-culture with methanogens
DOI:10.1111/j.1472-765X.1989.tb00323.x URL [本文引用: 1]
Dietary fiber future directions: integrating new definitions and findings to inform nutrition research and communication
DOI:10.3945/an.112.002907 URL [本文引用: 1]
Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin
A proposed taxonomy of anaerobic fungi (class Neocallimastigomycetes) suitable for large-scale sequence-based community structure analysis
DOI:10.1371/journal.pone.0036866 URL [本文引用: 1]
Towards a unified paradigm for sequence-based identification of fungi
DOI:10.1111/mec.12481 URL [本文引用: 1]
In-feed antibiotic effects on the swine intestinal microbiome
Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores
DOI:10.1038/ismej.2010.49 URL [本文引用: 1]
Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut
There is increasing interest in the human colonic microbiota and in the way its metabolic activities impact on host health and well-being. For most practical purposes, however, the large bowel is inaccessible for routine investigation, and a variety of animal and in vitro model systems have been developed to study the microbiota. In vitro models range from simple closed systems using pure or defined mixed populations of bacteria, or faecal material, to more sophisticated complex multistage continuous cultures that are able to simulate many of the spatial, temporal and environmental attributes that characterize microbiological events in different regions of the large gut. Recent developments using these systems have enabled modelling of surface colonisation and biofilm development, a hitherto neglected area of study.
Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals
DOI:10.1002/mbo3.2017.6.issue-5 URL [本文引用: 1]
Multivariate analyses in microbial ecology
Environmental microbiology is undergoing a dramatic revolution due to the increasing accumulation of biological information and contextual environmental parameters. This will not only enable a better identification of diversity patterns, but will also shed more light on the associated environmental conditions, spatial locations, and seasonal fluctuations, which could explain such patterns. Complex ecological questions may now be addressed using multivariate statistical analyses, which represent a vast potential of techniques that are still underexploited. Here, well-established exploratory and hypothesis-driven approaches are reviewed, so as to foster their addition to the microbial ecologist toolbox. Because such tools aim at reducing data set complexity, at identifying major patterns and putative causal factors, they will certainly find many applications in microbial ecology.
Diversity and occurrence of coprophilous fungi
DOI:10.1017/S0953756201003884 URL [本文引用: 1]
Records of coprophilous fungi from the Lesser Antilles and Puerto Rico
DOI:10.18475/cjos.v44i2.a8 URL [本文引用: 1]
Ribosomal ITS1 sequence- based diversity analysis of anaerobic rumen fungi in cattle fed on high fiber diet
DOI:10.1007/s13213-013-0620-2 URL [本文引用: 1]
Sporovexins AC and a new preussomerin analog: antibacterial and antifungal metabolites from the coprophilous fungus Sporormiella vexans
Sporovexins A-C (1-3) and 3'-O-desmethyl-1-epipreussomerin C (4) have been isolated from liquid cultures of the coprophilous fungus Sporormiella vexans (JS 306). The structures of these new metabolites were elucidated on the basis of MS and NMR analysis. Compounds 1 and 4 show antifungal activity against competitor fungi, as well as antibacterial effects.
Coprophilous fungi of the Falkland Islands
DOI:10.1017/S0960428610000156 URL [本文引用: 1]
The preussomerins: novel antifungal metabolites from the coprophilous fungus Preussia isomera Cain
DOI:10.1021/jo00014a007 URL [本文引用: 1]
The microbial community in the feces of the giant panda (Ailuropoda melanoleuca) as determined by PCR-TGGE profiling and clone library analysis
DOI:10.1007/s00248-007-9225-2 URL [本文引用: 1]
Human gut microbiome viewed across age and geography
DOI:10.1038/nature11053 URL [本文引用: 1]
Mitochondrial phylogeography and genetic diversity of Tibetan gazelle (Procapra picticaudata): implications for conservation
DOI:10.1016/j.ympev.2006.05.024 URL [本文引用: 1]
Impacts of grassland fence on the behavior and habitat area of the critically endangered Przewalski’s gazelle around the Qinghai Lake
DOI:10.1007/s11434-013-5844-9 URL [本文引用: 1]