 PDF(1037 KB)
PDF(1037 KB)


 PDF(1037 KB)
PDF(1037 KB)
 PDF(1037 KB)
PDF(1037 KB)
基于转录组分析高温抑制广东虫草原基形成的调控网络
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Dissecting the regulation network of high temperature inhibiting primordial formation of Cordyceps guangdongensis based on transcriptomic data
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}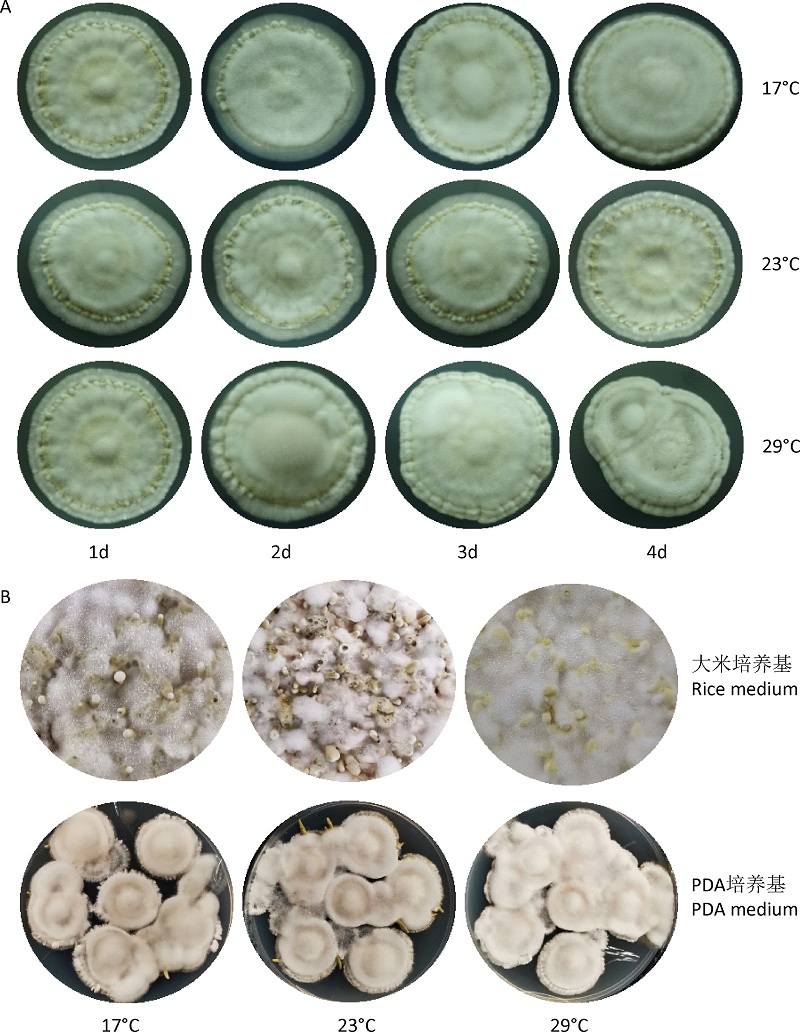
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |