 PDF(662 KB)
PDF(662 KB)


冬虫夏草菌实时荧光定量PCR内参基因的筛选
苏强军,夏樱霞,谢放,Uwitugabiye VESTINE,陈照禾,周刚
菌物学报 ›› 2021, Vol. 40 ›› Issue (7) : 1712-1722.
 PDF(662 KB)
PDF(662 KB)
 PDF(662 KB)
PDF(662 KB)
冬虫夏草菌实时荧光定量PCR内参基因的筛选
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Screening of the reference genes for qRT-PCR analysis of gene expression in Ophiocordyceps sinensis
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}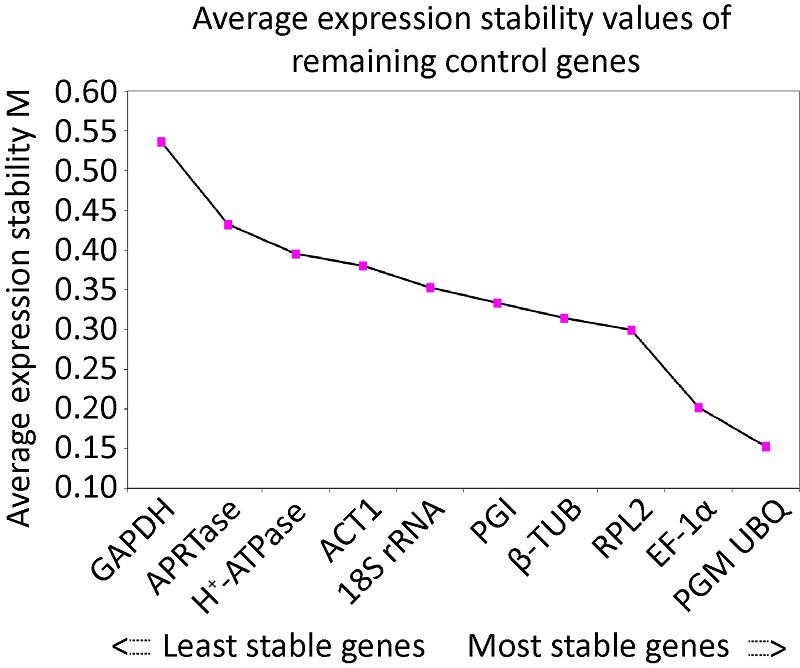
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |