新世纪以来,中国菌物学研究飞速发展,取得了令人瞩目的成就。在系统分类学方面,我国学者对担子菌门、“酵母菌”、蘑菇目、牛肝菌目、多孔菌目、亚隔孢壳科,以及一些具有广泛经济重要性的属如木霉属、刺盘孢属、青霉属、曲霉属等类群的系统发育构建和分类系统更新做出了重要贡献,并描述了大量的新分类单元(Zeng et al. 2013;Zhao et al. 2013;Liu et al. 2014;Wang et al. 2014;Wu et al. 2014;Wang et al. 2015;Zhao et al. 2016;Chen et al. 2017;Chen & Zhuang 2017;Zhao et al. 2017;Wang et al. 2018;Cui et al. 2019;He et al. 2019;Hou et al. 2020)。信息库建设方面,中国学者自主创建的菌物名称注册信息库(Fungal Names,
在大数据时代,生物多样性数据库中蕴藏着大量具有重要价值的生物学与生态学信息,如物种名称的变化、分类地位的改变、命名人、物种地理分布、生境、采集年代和时间等。这些信息不仅能揭示地区性物种多样性、生物资源调查研究的广度和深度,也可反映分类学及其相关学科的发展态势。本研究通过收集菌物名称发表数据,开展深入挖掘与分析,总结21世纪中国学者发表菌物新分类单元的情况和中国的菌物新物种发现概况,从而揭示中国菌物分类学的发展趋势。
1 数据来源
本研究的数据来自世界菌物名称信息库Index Fungorum(
根据Index Fungorum中的“Authors of fungal names”数据集,从原始数据中遴选出中国学者参与发表的记录共9 430条,用以分析中国学者在新分类单元发表方面的情况和贡献。根据分类阶元和模式标本产地的信息,从原始数据中筛选出中国菌物新物种发表记录共5 458条,作为中国菌物新物种的发表情况的分析数据。
2 中国学者发表菌物新名称概况
2.1 在世界的总体贡献
对菌物名称发表数据进行整体分析,发现2000-2020年间有1 491位中国学者参与了新分类单元及新名称的发表,发表了各类型菌物新名称9 430个,占全世界命名作者总数的14.31%和新名称总数的14.04%。中国学者参与发表的菌物新分类单元有7 120个,包括新纲3个、新目24个、新科88个、新亚科4个、新属492个、新亚属3个、新组及新亚组23个、新种6 404个和新种下单元(亚种、变种和变型)79个。此外,还发表了新组合1 868个、新修订名称61个和其他名称(不合格、不合法和弃用名称)381个。对各个名称的命名作者进行统计,其中56.11%(5 291个)的名称由中国作者独立完成发表,43.89%(4 139个)的名称与外国人合作发表。不同等级的新名称,中外作者合作的情况也不尽相同。新纲、新目和新科等高阶分类阶元,80%以上由中外作者合作发表,而60%以上新种及种下单元的发表由中国作者独立完成(表1)。
表1 2000-2020年中国学者发表的各类型菌物新分类单元及新名称
Table 1
| 名称类型 New taxa | 中国学者发表 Published by Chinese scholars | 中国学者独立发表 Published solely by Chinese scholars | 独立发表比例 Percentage of names solely published by Chinese scholars (%) |
|---|---|---|---|
| 新纲 New classes | 3 | 0 | 0.00 |
| 新目 New orders | 24 | 4 | 16.67 |
| 新科 New families | 88 | 18 | 20.45 |
| 新亚科 New subfamilies | 4 | 4 | 100.00 |
| 新属 New genera | 492 | 167 | 33.94 |
| 新亚属 New subgenera | 3 | 1 | 33.33 |
| 新组、新亚组 New sections and subsections | 23 | 12 | 52.17 |
| 新种 New species | 6 404 | 4 057 | 63.35 |
| 新种下单元 New infraspecific taxa | 79 | 53 | 67.09 |
| 新组合 New combinations | 1 868 | 759 | 40.63 |
| 名称修订 New revised names | 61 | 27 | 45.00 |
| 不合格、不合法名称 Invalid and illegitimate names | 381 | 189 | 49.61 |
| 总计 Total | 9 430 | 5 291 | 56.11 |
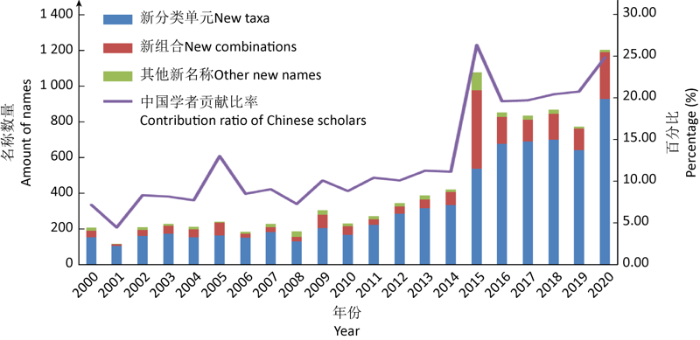
2000-2020年间,中国研究人员发表的菌物新名称数量快速增长,从事新名称发表的作者数量也在稳步上升。21世纪初,中国学者参与发表的名称仅占世界的5%-10%,而这一比例在2015年之后大幅提升。2015年以来,全世界共发表菌物新名称25 467个,中国学者参与了其中1/5以上的名称发表(图1)。中国是公认的世界生物多样性最丰富的国家之一,仍有大量菌物物种尚未被发现,随着研究队伍的稳定和逐步扩大,可以预见在未来一段时间内,我国将继续保持菌物分类学的高产出趋势。
图1
图1
2000-2020年中国学者发表菌物新名称数量及在世界的贡献
Fig. 1
New fungal names published by Chinese scholars and the contribution of Chinese scholars to the world from 2000 to 2020.
2.2 命名作者
表2 2000-2020年发表100个以上菌物新名称的中国学者
Table 2
| 命名作者 Author of new names | 新高阶分类单元 New higher taxa | 新种及种下单元 New species and infraspecific taxa | 新组合 New combinations | 其他新名称 Other new names | 总计 Total |
|---|---|---|---|---|---|
| 蔡磊 L. Cai | 37 | 463 | 157 | 18 | 675 |
| 戴玉成 Y.C. Dai | 44 | 391 | 177 | 10 | 622 |
| 白逢彦 F.Y. Bai | 81 | 216 | 248 | 72 | 617 |
| 杨祝良 Zhu L. Yang | 32 | 292 | 142 | 12 | 478 |
| 王启明 Q.M. Wang | 61 | 160 | 150 | 49 | 420 |
| 庄文颖 W.Y. Zhuang | 9 | 279 | 50 | 4 | 342 |
| 崔宝凯 B.K. Cui | 33 | 200 | 89 | 2 | 324 |
| 张修国 X.G. Zhang | 15 | 272 | 11 | 0 | 298 |
| 张天宇 T.Y. Zhang | 1 | 242 | 3 | 5 | 251 |
| 刘新展 Xin Zhan Liu | 23 | 23 | 102 | 29 | 177 |
| 朱宇敏 Y.M. Ju | 7 | 84 | 64 | 3 | 158 |
| 赵瑞琳 R.L. Zhao | 8 | 137 | 5 | 3 | 153 |
| 卢永仲 Y.Z. Lu | 19 | 87 | 41 | 5 | 152 |
| 陈倩 Qian Chen | 10 | 50 | 80 | 8 | 148 |
| 罗宗龙 Z.L. Luo | 14 | 120 | 5 | 2 | 141 |
| 李泰辉 T.H. Li | 4 | 128 | 5 | 4 | 141 |
| 田呈明 C.M. Tian | 15 | 107 | 7 | 3 | 132 |
| 刘芳 F. Liu | 8 | 105 | 15 | 1 | 129 |
| 周丽伟 L.W. Zhou | 10 | 85 | 31 | 2 | 128 |
| 吴声华 Sheng H. Wu | 9 | 76 | 37 | 3 | 125 |
| 何双辉 S.H. He | 7 | 76 | 35 | 2 | 120 |
| 张颖 Y. Zhang | 18 | 46 | 48 | 4 | 116 |
| 李爱华 A.H. Li | 0 | 110 | 1 | 1 | 112 |
| 袁海生 H.S. Yuan | 4 | 99 | 6 | 1 | 110 |
| 梁宗琦 Z.Q. Liang | 2 | 93 | 10 | 1 | 106 |
| 郭林 L. Guo | 1 | 103 | 1 | 0 | 105 |
| 范鑫磊 X.L. Fan | 8 | 63 | 29 | 2 | 102 |
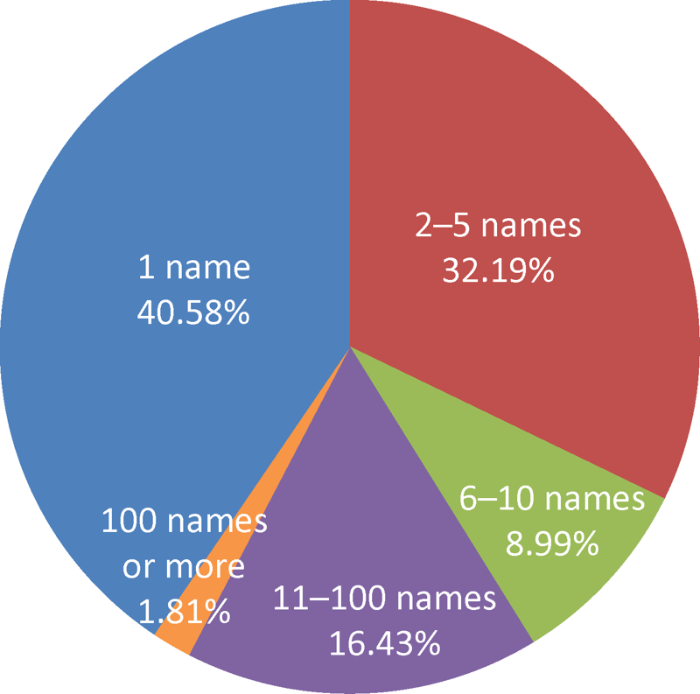
图2
图2
2000-2020年发表不同数量菌物新名称的中国学者占比
Fig. 2
Magnitude proportion of Chinese scholars who published new fungal names from 2000 to 2020.
2.3 研究类群
新世纪以来,中国学者的菌物研究类群涵盖真菌(true fungi)、卵菌(oomycetes)和黏菌(slime mold)3个类群,涉及11门43纲173目525科1 997属。根据中国菌物名录数据库(
以科为单位,对各个类群的研究情况进行分析,统计发现中国学者在近21年侧重于在大型木生真菌(刺革菌科和多孔菌科)、牛肝菌(牛肝菌科)、小型子囊菌(亚隔孢壳科、隔孢腔菌科、间座壳科、毛壳科)和伞菌(蘑菇科和红菇科)等类群开展研究(表3)。值得注意的是,中国学者发表的103个新属和709个新种在科等级的分类地位未定(Incertae sedis),占新属总数的1/5和新种总数的1/10,这说明菌物的高阶分类系统还远未澄清。
表3 2000-2020年中国学者发表新分类单元最多的20个科
Table 3
| 科 Family | 新属及亚属 New genera and subgenera | 新种及种下单元 New species and infraspecific taxa | 新组合 New combinations | 总数 Total |
|---|---|---|---|---|
| 刺革菌科 Hymenochaetaceae | 7 | 212 | 83 | 302 |
| 牛肝菌科 Boletaceae | 19 | 169 | 83 | 271 |
| 亚隔孢壳科 Didymellaceae | 17 | 119 | 102 | 238 |
| 多孔菌科 Polyporaceae | 17 | 158 | 49 | 224 |
| 格孢腔菌科 Pleosporaceae | 1 | 176 | 19 | 196 |
| 蘑菇科 Agaricaceae | 14 | 146 | 19 | 179 |
| 间座壳科 Diaporthaceae | 2 | 157 | 10 | 169 |
| 毛壳科 Chaetomiaceae | 18 | 78 | 79 | 175 |
| 炭角菌科 Xylariaceae | 11 | 128 | 29 | 168 |
| 毛筒壳科 Tubeufiaceae | 20 | 102 | 44 | 166 |
| 红菇科 Russulaceae | 6 | 135 | 6 | 147 |
| 球囊菌科 Mycosphaerellaceae | 3 | 98 | 19 | 120 |
| 丛赤壳科 Nectriaceae | 4 | 106 | 19 | 129 |
| 肉座菌科 Hypocreaceae | 0 | 117 | 6 | 123 |
| 斑痣盘菌科 Rhytismataceae | 1 | 112 | 5 | 118 |
| 圆盘菌科 Orbiliaceae | 3 | 68 | 29 | 100 |
| 拟盘多毛孢科 Pestalotiopsidaceae | 0 | 69 | 22 | 91 |
| 布勒担菌科 Bulleribasidiaceae | 5 | 51 | 33 | 89 |
| 曲霉科 Aspergillaceae | 1 | 91 | 2 | 94 |
| 鹅膏科 Amanitaceae | 1 | 83 | 4 | 88 |
2.4 研究质量
在中国学者近21年发表的新名称中,有9 049个符合《国际藻类、菌物和植物命名法规》的各项条款,属于正确发表名称,占名称总数的95.96%。其余381个新名称中,不合法名称有52个、不合格名称有329个,这些名称所违反的法规条款和主要存在问题见表4。其中,模式指定是我国学者在发表菌物新名称时出现问题最多的部分。建议在今后的工作中,严格遵守《国际藻类、菌物和植物命名法规》,将模式标本保藏在标本馆,尤其是条件优良管理规范的标本馆等保藏机构,并在撰写文章时正确指明标本号和“typus”等注释语,进一步提升名称发表的质量。
表4 2000-2020年中国学者不合格或不合法发表名称的类型和数量
Table 4
| 名称类型 Type of names | 违反的命名法规条款 Violated articles in code | 主要存在的问题 Major problems | 名称数量 Number of names |
|---|---|---|---|
| 不合法 Illegitimate name | 52.1 | 命名多余名称 Nomenclaturally superfluous names | 12 |
| 不合法 Illegitimate name | 53.1 | 晚出同名,与更早发表的名称拼写完全相同 Later homonym | 40 |
| 不合格 Invalid name | 38.1 | 没有写出或引证描述和特征集要 New taxon published without a description or diagnosis | 10 |
| 不合格 Invalid name | 39.1 | 在2011年12月31日前发表的名称,没有写出或引证拉丁文描述和特征集要 Before 31 December 2011, a new taxon is published without a Latin description or diagnosis | 25 |
| 不合格 Invalid name | 40.1 | 属或以下等级的新分类群名称,没有指明命名模式 The type is not indicated when a new taxon at the rank of genus or below is published | 32 |
| 不合格 Invalid name | 40.6 | 属或以下等级的新分类群名称,指定模式时没有注明“typus[模式]”或“holotypus[主模式]”、或其同义等同语 One of the words “typus” or “holotypus”, or its equivalent in a modern language is not included when a new taxon at the rank of genus or below is published | 24 |
| 不合格 Invalid name | 40.7 | 新种或种下分类群名称,没有指明或指出了多于1个模式保存的标本馆或收藏机构 The single herbarium or conserved institution in which the type is conserved is not specified when a new species or infraspecific taxon is published | 108 |
| 不合格 Invalid name | 41.5 | 发表新组合时,没有完整的引证其基名或被替代异名 The basionym or replaced synonym is not clearly indicated when a new combination is published | 63 |
| 不合格 Invalid name | F.5.1 | 没有引证名称注册号 Without citation of an identifier issued by a recognized repository for the new name | 25 |
| 不合格 Invalid name | 其他不合格发表条款 Other articles involving invalidly published names | 42 |
2.5 发表来源
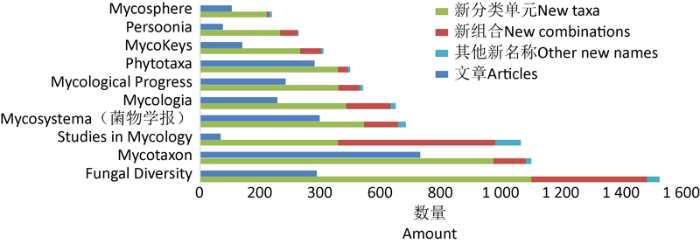
新世纪以来,中国学者将9 430个菌物名称发表在205个期刊和书籍等出版物中。其中9 176个名称发表在169个期刊的4 029篇文章中,占新名称总数的97.31%;剩余254个新名称发表在36部专著、志书等著作中。在出版物的选择上,中国学者更倾向于在英文期刊发表新分类单元及新名称,90%以上的新名称和相关文章发表在136个英文期刊上。其中,《Fungal Diversity》杂志刊载了最多的新名称(1 530个)和新高阶分类单元(151个),《Mycotaxon》杂志上发表了最多数量的文章(734篇)和新物种(945个),而《Studies in Mycology》是发表新组合(524个)最多的期刊(图3)。除此之外,仅有8.92%的新名称(841个)和12.11%(488篇)的文章发表在中文期刊上。在这些名称中,有81.45%(685个)发表在《菌物学报》上,剩余156个名称分别发表在《菌物研究》《云南植物研究》(即现在的《Plant Diversity》)、《广西农业生物科学》等32个中文期刊的89篇文章中。
图3
图3
2000-2020年中国学者发表菌物新名称最多的10个期刊
Fig. 3
Top 10 journals in which Chinese scholars have published new fungal names from 2000 to 2020.
3 中国的菌物新物种发现
3.1 新物种发表情况
2000年到2020年间,全世界共发表菌物新物种35 436种,其中来自中国的新物种有5 458种,占15.40%。这些新物种中,由中国学者独立完成发表的有3 890个,占71.27%;由中外学者合作发表的有1 048个,占19.20%;剩余520个由外国学者发表,占9.53%。
3.2 新物种的地域分布
表5 2000-2020年中国各省发现的菌物新物种数量
Table 5
| 省 Province | 新物种数量 Number of new species | 省 Province | 新物种数量 Number of new species | 省 Province | 新物种数量 Number of new species |
|---|---|---|---|---|---|
| 云南 Yunnan | 1 345 | 江西 Jiangxi | 106 | 甘肃 Gansu | 63 |
| 海南 Hainan | 378 | 湖南 Hunan | 105 | 辽宁 Liaoning | 61 |
| 台湾 Taiwan | 355 | 陕西 Shaanxi | 105 | 山东 Shandong | 60 |
| 广东 Guangdong | 320 | 北京 Beijing | 105 | 河北 Hebei | 33 |
| 广西 Guangxi | 275 | 新疆 Xinjiang | 101 | 江苏 Jiangsu | 31 |
| 贵州 Guizhou | 249 | 安徽 Anhui | 97 | 山西 Shanxi | 31 |
| 四川 Sichuan | 229 | 青海 Qinghai | 85 | 宁夏 Ningxia | 23 |
| 西藏 Xizang | 208 | 黑龙江 Heilongjiang | 77 | 重庆 Chongqing | 17 |
| 湖北 Hubei | 148 | 浙江 Zhejiang | 71 | 上海 Shanghai | 1 |
| 香港 Hong Kong | 123 | 内蒙古 Inner Mongolia | 64 | 天津 Tianjin | 1 |
| 福建 Fujian | 113 | 河南 Henan | 63 | 澳门 Macao | 0 |
| 吉林 Jilin | 113 |
3.3 新物种的基质和寄主
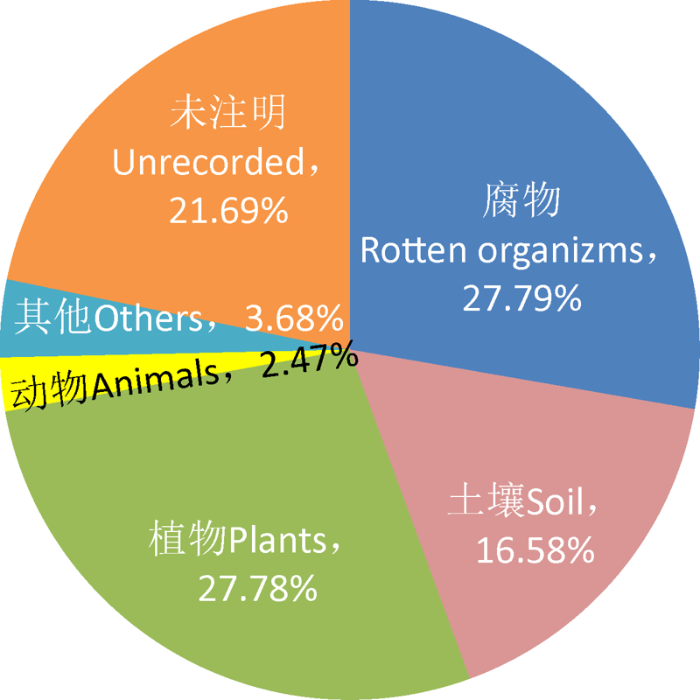
在中国发表的5 458个新物种中,注明生长基质或寄主的有4 274种,其中生长在腐木、落叶、粪便、腐殖质等腐物以及活体植物上的真菌最多,分别有1 517种和1 516种,各占新物种总数的1/4以上。其次,有905种生长或分离自土壤中,占16.58%;剩余335种寄生在动物身上或生长在岩石、水等基物中(图4)。
图4
图4
2000-2020年中国菌物新物种的来源基质和/或寄主
Fig. 4
Substrate and/or host origins of new fungal species from China published from 2000 to 2020.
4 展望
21世纪以来,我国菌物分类学研究类群逐渐扩大、研究人员稳步增加,取得了丰硕的研究成果,在世界范围也做出了突出的贡献并得到广泛认可。展望下一个20年,菌物分类学仍有许多重大问题尚未解决。首先,我国尚未完成菌物资源的全面普查,仍有许多地区的菌物多样性和资源分布信息处于空白状态,亟需开展相应的研究工作。据保守估计,全球菌物物种多样性有220-380万种(Hawksworth & Lücking 2017),而目前仅发现了不到6%,其描述率为地球主要生物类群中最低,仍有大量菌物新物种有待发现。我国菌物多样性也十分丰富,初步估计有超过30万种(魏江春 2010),已有一些研究初步统计出我国已知菌物物种约为1.25-2.79万种之间(戴玉成和庄剑云 2010;Fang et al. 2018; The Biodiversity Committee of Chinese Academy of Sciences 2020),目前已发现的物种仅有4%-9%。再者,菌物资源的开发应用范围需进一步扩大。全世界已经广泛认可菌类是人类最佳饮食结构的组成部分之一,而研究表明多种菌物具有多糖、核苷、微量元素等多种有益成分,可用于免疫调节、抗病毒、抑肿瘤等功效。目前,中国已报道的食药用菌数量为世界最多,有2 026种,而可人工栽培的仅有100余种,仍有大量具有开发潜质的物种(Fang et al. 2018)。除此之外,菌物可在生态、工业、农业等多个领域发挥效用,在现有工作基础上,进一步拓宽应用范围可以更充分地发挥菌物资源的价值。我国菌物资源的保护工作仍处于起步状态,亟须全面展开。目前全国仅有吉林天佛指山国家级自然保护区对野生松口蘑资源进行了保护,对其他受威胁物种还未采取有效的保护政策和措施。在今后的工作中需要填补对菌物资源野外检测的空白,同时在政策制定、就地保护、离体保存、科学普及等全方面开展保护工作,以促进菌物资源的可持续利用。
致谢
姚一建研究员、马克平研究员对本文部分内容给予过建议,赵明君女士提供过数据库相关技术协助,特致以谢意。
参考文献
Discovery of a large-scaled survey of Trichoderma in soil from China
DOI:10.1038/s41598-017-07807-3
PMID:28831112
[本文引用: 1]

The first large-scaled survey of soil-inhabiting Trichoderma is conducted in 23 provinces of China. Twenty-three new species belonging to the green-ascospored clades are discovered. Their phylogenetic positions are determined by sequence analyses of the combined partial sequences of translation elongation factor 1-alpha and the second largest RNA polymerase subunit encoding genes. Morphology and culture characteristics are observed, described and illustrated in detail. Distinctions between the new species and their close relatives are compared and discussed. They are named as: T. aggregatum, T. alpinum, T. bannaense, T. breve, T. brevicrassum, T. byssinum, T. chlamydosporicum, T. concentricum, T. ganodermatis, T. hainanense, T. hengshanicum, T. hirsutum, T. hunanense, T. ingratum, T. liberatum, T. linzhiense, T. longisporum, T. polypori, T. pseudodensum, T. simplex, T. solum, T. undatipile and T. zayuense.
Didymellaceae revisited
DOI:10.1016/j.simyco.2017.06.002 URL [本文引用: 1]
Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China
DOI:10.1007/s13225-019-00427-4 URL [本文引用: 1]
Diversity and systematics of the important macrofungi in Chinese forests
Numbers of fungal species hitherto known in China
Publications dealing with Chinese fungal species were systematically investigated, and 2,849 new species, 129 new varieties and 5,260 new Chinese records have been reported during 1978 to 2010. “Sylloge Fungorum Sinicorum” was published in 1979, listed 6,737 species and 168 varieties based on the reports on Chinese fungal resource until 1973. So 14,846 species and 297 varieties have been found in mainland China by 2010. In addition, 2,122 fungal species were recorded in Hong Kong and 6,207 in Taiwan, among which around 800 and 400 species, respectively, were not found in mainland China so far. Until now there are 16,046 species and 297 varieties have been recorded in the Chinese territory. If 10% of them are the synonyms, the Chinese fungal species are around 14,700. Among them around 300 species are Chromista, 340 are Protoza, and 14,060 are Fungi.
Country focus: China
Fungal diversity revisited: 2.2 to 3.8 million species
Notes, outline and divergence times of Basidiomycota
DOI:10.1007/s13225-019-00435-4 URL [本文引用: 1]
The phoma-like dilemma
DOI:10.1016/j.simyco.2020.05.001 URL [本文引用: 1]
The Colletotrichum gigasporum species complex
DOI:10.3767/003158514X684447
URL
[本文引用: 1]

In a preliminary analysis, 21 Colletotrichum strains with large conidia preserved in the CBS culture collection clustered with a recently described species, C. gigasporum, forming a clade distinct from other currently known Colletotrichum species complexes. Multi-locus phylogenetic analyses (ITS, ACT, TUB2, CHS-1, GAPDH) as well as each of the single-locus analyses resolved seven distinct species, one of them being C. gigasporum. Colletotrichum gigasporum and its close allies thus constitute a previously unknown species complex with shared morphological features. Five of the seven species accepted in the C. gigasporum species complex are described here as novel species, namely C. arxii, C. magnisporum, C. pseudomajus, C. radicis and C. vietnamense. A species represented by a single sterile strain, namely CBS 159.50, was not described as novel species, and is treated as Colletotrichum sp. CBS 159.50. Furthermore, C. thailandicum is reduced to synonymy with C. gigasporum.
Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina
DOI:10.3767/003158514X682313
URL
[本文引用: 1]

Ustilaginomycotina (Basidiomycota, Fungi) has been reclassified recently based on multiple gene sequence analyses. However, the phylogenetic placement of two yeast-like genera Malassezia and Moniliella in the subphylum remains unclear. Phylogenetic analyses using different algorithms based on the sequences of six genes, including the small subunit (18S) ribosomal DNA (rDNA), the large subunit (26S) rDNA D1/D2 domains, the internal transcribed spacer regions (ITS 1 and 2) including 5.8S rDNA, the two subunits of RNA polymerase II (RPB1 and RPB2) and the translation elongation factor 1-alpha (EF1-alpha), were performed to address their phylogenetic positions. Our analyses indicated that Malassezia and Moniliella represented two deeply rooted lineages within Ustilaginomycotina and have a sister relationship to both Ustilaginomycetes and Exobasidiomycetes. Those clades are described here as new classes, namely Moniliellomycetes with order Moniliellales, family Moniliellaceae, and genus Moniliella; and Malasseziomycetes with order Malasseziales, family Malasseziaceae, and genus Malassezia. Phenotypic differences support this classification suggesting widely different life styles among the mainly plant pathogenic Ustilaginomycotina.
Phylogenetic classification of yeasts and related taxa within Pucciniomycotina
DOI:10.1016/j.simyco.2015.12.002 URL [本文引用: 1]
Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species
DOI:10.1038/s41598-016-0028-x URL [本文引用: 1]
The biodiversity of pan-fungi and the sustainable development of human beings
Molecular phylogenetic analyses redefine seven major clades and reveal 22 new generic clades in the fungal family Boletaceae
DOI:10.1007/s13225-014-0283-8
URL
[本文引用: 1]

Mushrooms in the basidiomycete family Boletaceae are ecologically and economically very important. However, due to the morphological complexity and the limited phylogenetic information on the various species and genera of this fungal family, our understanding of its systematics and evolution remains rudimentary. In this study, DNA sequences of four genes (nrLSU, tef1-alpha, rpb1, and rpb2) were newly obtained from ca. 200 representative specimens of Boletaceae. Our phylogenetic analyses revealed seven major clades at the subfamily level, namely Austroboletoideae, Boletoideae, Chalciporoideae, Leccinoideae, Xerocomoideae, Zangioideae, and the Pulveroboletus Group. In addition, 59 genus-level clades were identified, of which 22 were uncovered for the first time. These 22 clades were mainly placed in Boletoideae and the Pulveroboletus Group. The results further indicated that the characters frequently used in the morphology-based taxonomy of Boletaceae, such as basidiospore ornamentation, the form of the basidioma, and the stuffed pores each had multiple origins within the family, suggesting that the use of such features for high-level classification of Boletaceae should be de-emphasized and combined with other characters.
Development of red list assessment of macrofungi in China
DOI:10.17520/biods.2019173 URL
The genus Phylloporus (Boletaceae, Boletales) from China: morphological and multilocus DNA sequence inference
DOI:10.1007/s13225-012-0184-7
URL
[本文引用: 1]

Species of the genus Phylloporus in China were investigated based on morphology and molecular phylogenetic analysis of a three-locus (nrLSU, ITS and tef-1a) DNA sequence dataset. Twenty-one phylogenetic species were recognized among the studied collections. Seven of them are described as new: P. brunneiceps, P. imbricatus, P. maculatus, P. pachycystidiatus, P. rubeolus, P. rubrosquamosus, and P. yunnanensis. In addition, four of them correspond with the previous morphology-based taxa: P. bellus, P. luxiensis, P. parvisporus, and P. rufescens. The remaining ten phylogenetic species were not described due to the paucity of the materials. A key to the Chinese morphologically recognizable taxa was provided. A preliminary biogeographical analysis showed that (1) Pylloporus species in East Asia and Southeast Asia are mostly closely related, (2) species pairs or closely related species of Phylloporus between East Asia and North/Central America are relatively common, and (3) the biogeographic relationship of Phylloporus between East Asia and Europe was supported by only a single species pair. Unexpectedly, no taxa common either to both Europe and East Asia, or to both East Asia and North/Central America, were uncovered. Clades look to have taxa from both sides of the Pacific and Europe/Asia though.
New species and phylogeny of Perenniporia based on morphological and molecular characters
DOI:10.1007/s13225-012-0177-6
URL
[本文引用: 1]

Three new resupinate, poroid, wood-inhabiting fungi, Perenniporia aridula, P. bannaensis and P. substraminea, are introduced on the basis of morphological and molecular characters. Molecular study based on sequence data from the ribosomal ITS and LSU regions supported the three new species' positions in Perenniporia s.s., and all of them formed monophyletic lineages with strong support (100 % BP, 1.00 BPP). Phylogenetic analysis revealed seven clades for the 31 species of Perenniporia s.l. used in this study. Among them, Perenniporiella clustered with Perenniporia ochroleuca group, and then subsequently grouped with Abundisporus. In addition, the P. ochroleuca group, the P. vicina group, the P. martia group and P. subacida formed well supported monophyletic entities, which could be recognized as distinct genera, and they are not related to P. medulla-panis which belongs to Perenniporia s.s. clade. An identification key to 38 species of Perenniporia occurring in China is provided.
A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective
DOI:10.1007/s13225-017-0381-5 URL [本文引用: 1]
Towards standardizing taxonomic ranks using divergence times - a case study for reconstruction of the Agaricus taxonomic system
DOI:10.1007/s13225-016-0357-x URL [本文引用: 1]
中国森林大型真菌重要类群多样性和系统学研究
中国菌物已知种数
据作者对1978年以来国内外主要菌物学期刊和专著进行的系统搜集,中国大陆发表的菌物累计有2,849新种,129新变种,5,260新记录种。若加上戴芳澜编著的《中国真菌总汇》所记载的6,737种和168变种,中国大陆已知菌物计14,846种297变种。据不完全统计,香港和台湾报道的菌物种类中分别约有800种和400种在中国大陆未曾记载,因此全国总计已知菌物应为16,046种297变种。假设其中有10%为同物异名,则目前我国菌物已知种数约为14,700种,其中管毛生物界(主要是卵菌)约300种,原生动物界(主要是黏菌)约有340种,真菌约14,060种。
中国大型真菌红色名录评估研究进展
DOI:10.17520/biods.2019173
PMID:EAD036E2-0221-4F52-B650-80CF1E8509A7
[本文引用: 1]

大型真菌具有重要的生态价值和经济价值, 但由于环境污染、气候变化、生境丧失与破碎化, 以及资源过度利用等因素, 其生物多样性受到严重威胁。为了全面评估中国大型真菌的生存状况, 国家生态环境部(原环境保护部)联合中国科学院于2016年启动了《中国生物多样性红色名录——大型真菌卷》的编制工作。经广泛和全面收集文献资料, 依据IUCN物种红色名录等级与标准, 结合大型真菌特点和国内研究现状, 制定了中国大型真菌红色名录评估方法和流程, 动员和组织了全国相关研究力量, 对9,302种大型真菌的受威胁状况进行了评估。结果显示, 中国大型真菌受威胁物种(包括疑似灭绝、极危、濒危、易危)共97个, 占被评估物种总数的1.04%; 近危101种, 占总数的1.09%; 无危2,764种, 占总数的29.71%; 数据不足6,340种, 占总数的68.16%。此次评估工作汇集了全国140多位专家的智慧, 是国内外迄今为止涉及物种数量最大、类群范围最宽、覆盖地域最广、参与人员最多的一次大型真菌生存状况评估, 对我国大型真菌多样性保护与管理具有重要意义。